Circulating mitochondrial genes detect acute cardiac allograft rejection: Role of the mitochondrial calcium uniporter complex
- PMID: 33125788
- PMCID: PMC8246899
- DOI: 10.1111/ajt.16387
Circulating mitochondrial genes detect acute cardiac allograft rejection: Role of the mitochondrial calcium uniporter complex
Abstract
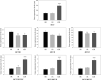
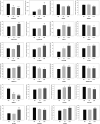
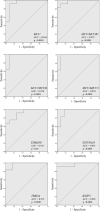
Acute rejection after heart transplantation increases the risk of chronic dysfunction. Disturbances in mitochondrial function may play a contributory role, however, the relationship between histological signs of rejection in the human transplanted heart and expression levels of circulating mitochondrial genes, such as the mitochondrial Ca2+ uniporter (MCU) complex, remains unexplored. We conducted an RNA-sequencing analysis to identify altered mitochondrial genes in serum and to evaluate their diagnostic accuracy for rejection episodes. We included 40 consecutive samples from transplant recipients undergoing routine endomyocardial biopsies. In total, 112 mitochondrial genes were identified in the serum of posttransplant patients, of which 28 were differentially expressed in patients with acute rejection (p < .05). Considering the receiver operating characteristic analysis with an area under the curve (AUC) >0.900 to discriminate patients with moderate or severe degrees of rejection, we found that the MCU system showed a strong capability for detection: MCU (AUC = 0.944, p < .0001), MCU/MCUR1 ratio (AUC = 0.972, p < .0001), MCU/MCUB ratio (AUC = 0.970, p < .0001), and MCU/MICU1 ratio (AUC = 0.970, p < .0001). Mitochondrial alterations are reflected in peripheral blood and are capable of discriminating between patients with allograft rejection and those not experiencing rejection with excellent accuracy. The dysregulation of the MCU complex was found to be the most relevant finding.
Keywords: biomarker; cell death: apoptosis; genomics; heart biology; heart transplantation/cardiology; molecular biology; rejection: acute; translational research/science.
© 2020 The Authors. American Journal of Transplantation published by Wiley Periodicals LLC on behalf of The American Society of Transplantation and the American Society of Transplant Surgeons.
Conflict of interest statement
The authors of this manuscript have no conflicts of interest to disclose as described by the
Figures

Comment in
-
Transcriptomics in transplantation: More than just biomarkers of allograft rejection.Am J Transplant. 2021 Jun;21(6):2000-2001. doi: 10.1111/ajt.16429. Epub 2020 Dec 21. Am J Transplant. 2021. PMID: 33278854 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

