Relative Levels of Gli1 and Gli2 Determine the Response of Ventral Neural Stem Cells to Demyelination
- PMID: 33125874
- PMCID: PMC7664046
- DOI: 10.1016/j.stemcr.2020.10.003
Relative Levels of Gli1 and Gli2 Determine the Response of Ventral Neural Stem Cells to Demyelination
Abstract
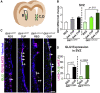
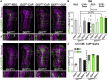
Enhancing repair of myelin is an important therapeutic goal in many neurological disorders characterized by demyelination. In the healthy adult brain, ventral neural stem cells (vNSCs) in the subventricular zone, marked by GLI1 expression, do not generate oligodendrocytes. However, in response to demyelination, their progeny are recruited to lesions where they differentiate into oligodendrocytes and ablation of GLI1 further enhances remyelination. GLI1 and GLI2 are closely related transcriptional activators but the role of GLI2 in remyelination by vNSCs is not clear. Here, we show that genetic ablation of Gli1 in vNSCs increases GLI2 expression and combined loss of both transcription factors decreases the recruitment and differentiation of their progeny in demyelinated lesions. These results indicate that GLI1 and GLI2 have distinct, non-redundant functions in vNSCs and their relative levels play an essential role in the response to demyelination.
Keywords: GLI1; GLI2; Gli1 inhibition; Gli2 inhibition; adult subventricular zone; neural stem cell; remyelination.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Ahn S., Joyner A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. - PubMed
-
- Bai C.B., Auerbach W., Lee J.S., Stephen D., Joyner A.L. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. - PubMed
-
- Brewster R., Mullor J.L., Ruiz i Altaba A. Gli2 functions in FGF signaling during antero-posterior patterning. Development. 2000;127:4395–4405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

