A Founder Effect Led Early SARS-CoV-2 Transmission in Spain
- PMID: 33127745
- PMCID: PMC7925114
- DOI: 10.1128/JVI.01583-20
A Founder Effect Led Early SARS-CoV-2 Transmission in Spain
Abstract
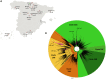
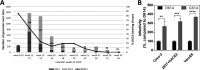
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) whole-genome analysis has identified five large clades worldwide which emerged in 2019 (19A and 19B) and in 2020 (20A, 20B, and 20C). This study aimed to analyze the diffusion of SARS-CoV-2 in Spain using maximum-likelihood phylogenetic and Bayesian phylodynamic analyses. The most recent common ancestor (MRCA) of the SARS-CoV-2 pandemic was estimated to have emerged in Wuhan, China, around 24 November 2019. Phylogenetic analyses of the first 12,511 SARS-CoV-2 whole-genome sequences obtained worldwide, including 290 from 11 different regions of Spain, revealed 62 independent introductions of the virus in the country. Most sequences from Spain were distributed in clades characterized by a D614G substitution in the S gene (20A, 20B, and 20C) and an L84S substitution in ORF8 (19B) with 163 and 118 sequences, respectively, with the remaining sequences branching in 19A. A total of 110 (38%) sequences from Spain grouped in four different monophyletic clusters of clade 20A (20A-Sp1 and 20A-Sp2) and 19B clade (19B-Sp1 and 19B-Sp2) along with sequences from 29 countries worldwide. The MRCAs of clusters 19A-Sp1, 20A-Sp1, 19A-Sp2, and 20A-Sp2 were estimated to have occurred in Spain around 21 and 29 January and 6 and 17 February 2020, respectively. The prevalence of clade 19B in Spain (40%) was by far higher than in any other European country during the first weeks of the epidemic, probably as a result of a founder effect. However, this variant was replaced by G614-bearing viruses in April. In vitro assays showed an enhanced infectivity of pseudotyped virions displaying the G614 substitution compared with those having D614, suggesting a fitness advantage of D614G.IMPORTANCE Multiple SARS-CoV-2 introductions have been detected in Spain, and at least four resulted in the emergence of locally transmitted clusters that originated not later than mid-February, with further dissemination to many other countries around the world, and a few weeks before the explosion of COVID-19 cases detected in Spain during the first week of March. The majority of the earliest variants detected in Spain branched in the clade 19B (D614 viruses), which was the most prevalent clade during the first weeks of March, pointing to a founder effect. However, from mid-March to June 2020, G614-bearing viruses (clades 20A, 20B, and 20C) overcame D614 variants in Spain, probably as a consequence of an evolutionary advantage of this substitution in the spike protein. A higher infectivity of G614-bearing viruses than D614 variants was detected, suggesting that this substitution in SARS-CoV-2 spike protein could be behind the variant shift observed in Spain.
Keywords: COVID-19; Europe; SARS-CoV-2; Spain; phylodynamics; phylogeography.
Copyright © 2021 Díez-Fuertes et al.
Figures





References
-
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- WHO. 2020. WHO Director-General's opening remarks at the media briefing on COVID-19, 11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-re.... Accessed 20 July 2020.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

