Netrin G1 Promotes Pancreatic Tumorigenesis through Cancer-Associated Fibroblast-Driven Nutritional Support and Immunosuppression
- PMID: 33127842
- PMCID: PMC7858242
- DOI: 10.1158/2159-8290.CD-20-0775
Netrin G1 Promotes Pancreatic Tumorigenesis through Cancer-Associated Fibroblast-Driven Nutritional Support and Immunosuppression
Abstract
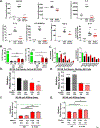
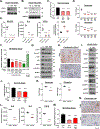
Pancreatic ductal adenocarcinoma (PDAC) has a poor 5-year survival rate and lacks effective therapeutics. Therefore, it is of paramount importance to identify new targets. Using multiplex data from patient tissue, three-dimensional coculturing in vitro assays, and orthotopic murine models, we identified Netrin G1 (NetG1) as a promoter of PDAC tumorigenesis. We found that NetG1+ cancer-associated fibroblasts (CAF) support PDAC survival, through a NetG1-mediated effect on glutamate/glutamine metabolism. Also, NetG1+ CAFs are intrinsically immunosuppressive and inhibit natural killer cell-mediated killing of tumor cells. These protumor functions are controlled by a signaling circuit downstream of NetG1, which is comprised of AKT/4E-BP1, p38/FRA1, vesicular glutamate transporter 1, and glutamine synthetase. Finally, blocking NetG1 with a neutralizing antibody stunts in vivo tumorigenesis, suggesting NetG1 as potential target in PDAC. SIGNIFICANCE: This study demonstrates the feasibility of targeting a fibroblastic protein, NetG1, which can limit PDAC tumorigenesis in vivo by reverting the protumorigenic properties of CAFs. Moreover, inhibition of metabolic proteins in CAFs altered their immunosuppressive capacity, linking metabolism with immunomodulatory function.See related commentary by Sherman, p. 230.This article is highlighted in the In This Issue feature, p. 211.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures





Comment in
-
A Presynaptic Protein Is a Net Gain for Pancreatic Tumor Progression.Cancer Discov. 2021 Feb;11(2):230-232. doi: 10.1158/2159-8290.CD-20-1645. Cancer Discov. 2021. PMID: 33531426
References
-
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, and Matrisian LM, Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res, 2014; 74: p. 2913–21. - PubMed
-
- Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2020. CA Cancer J Clin, 2020; 70: p. 7–30. - PubMed
-
- Ryan DP, Hong TS, and Bardeesy N, Pancreatic adenocarcinoma. N Engl J Med, 2014; 371: p. 2140–1. - PubMed
-
- Hidalgo M, Pancreatic cancer. N Engl J Med, 2010; 362: p. 1605–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 CA234221/CA/NCI NIH HHS/United States
- R21 CA231252/CA/NCI NIH HHS/United States
- R03 CA212949/CA/NCI NIH HHS/United States
- R01 CA168653/CA/NCI NIH HHS/United States
- R01 CA188430/CA/NCI NIH HHS/United States
- R01 CA113451/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- R01 CA232256/CA/NCI NIH HHS/United States
- R01 GM116911/GM/NIGMS NIH HHS/United States
- R01 CA194263/CA/NCI NIH HHS/United States
- P30 CA006927/CA/NCI NIH HHS/United States
- T32 CA009035/CA/NCI NIH HHS/United States
- 20-0081/AICR_/Worldwide Cancer Research/United Kingdom
- F32 CA213810/CA/NCI NIH HHS/United States
- R01 CA201276/CA/NCI NIH HHS/United States
- R35 CA242379/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

