Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics
- PMID: 33127901
- PMCID: PMC7603341
- DOI: 10.1038/s41522-020-00155-7
Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics
Abstract
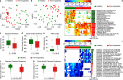
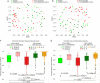
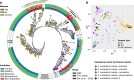
Dental implants are installed in an increasing number of patients. Mucositis and peri-implantitis are common microbial-biofilm-associated diseases affecting the tissues that surround the dental implant and are a major medical and socioeconomic burden. By metagenomic sequencing of the plaque microbiome in different peri-implant health and disease conditions (113 samples from 72 individuals), we found microbial signatures for peri-implantitis and mucositis and defined the peri-implantitis-related complex (PiRC) composed by the 7 most discriminative bacteria. The peri-implantitis microbiome is site specific as contralateral healthy sites resembled more the microbiome of healthy implants, while mucositis was specifically enriched for Fusobacterium nucleatum acting as a keystone colonizer. Microbiome-based machine learning showed high diagnostic and prognostic power for peri-implant diseases and strain-level profiling identified a previously uncharacterized subspecies of F. nucleatum to be particularly associated with disease. Altogether, we associated the plaque microbiome with peri-implant diseases and identified microbial signatures of disease severity.
Conflict of interest statement
P.G. and N.S. are co-founders of PreBiomics S.r.l., a company active in the fields of implantology and microbiome research. M.B. is an employee of PreBiomics, and C.T. is a scientific advisor for PreBiomics.
Figures


References
-
- Darcey J, Eldridge D. Fifty years of dental implant development: a continuous evolution. Dent. Hist. 2016;61:75–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

