Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR
- PMID: 33127953
- PMCID: PMC7599326
- DOI: 10.1038/s41598-020-75958-x
Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR
Abstract
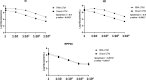
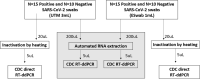
Droplet digital PCR (ddPCR) is a sensitive and reproducible technology widely used for quantitation of several viruses. The aim of this study was to evaluate the 2019-nCoV CDC ddPCR Triplex Probe Assay (BioRad) performance, comparing the direct quantitation of SARS-CoV-2 on nasopharyngeal swab with the procedure applied to the extracted RNA. Moreover, two widely used swab types were compared (UTM 3 mL and ESwab 1 mL, COPAN). A total of 50 nasopharyngeal swabs (n = 25 UTM 3 mL and n = 25 ESwab 1 mL) from SARS-CoV-2 patients, collected during the pandemic at IRCCS Sacro Cuore Don Calabria Hospital (Veneto Region, North-East Italy), were used for our purpose. After heat inactivation, an aliquot of swab medium was used for the direct quantitation. Then, we compared the direct method with the quantitation performed on the RNA purified from nasopharyngeal swab by automated extraction. We observed that the direct approach achieved generally equal RNA copies compared to the extracted RNA. The results with the direct quantitation were more accurate on ESwab with a sensitivity of 93.33% [95% CI, 68.05 to 99.83] and specificity of 100.00% for both N1 and N2. On the other hand, on UTM we observed a higher rate of discordant results for N1 and N2. The human internal amplification control (RPP30) showed 100% of both sensitivity and specificity independent of swabs and approaches. In conclusion, we described a direct quantitation of SARS-CoV-2 in nasopharyngeal swab. Our approach resulted in an efficient quantitation, without automated RNA extraction and purification. However, special care needs to be taken on the potential bias due to the conservation of samples and to the heating treatment, as we used thawed and heat inactivated material. Further studies on a larger cohort of samples are warranted to evaluate the clinical value of this direct approach.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Monleau M, et al. Evaluation of different RNA extraction methods and storage conditions of dried plasma or blood spots for human immunodeficiency virus type 1 RNA quantification and PCR amplification for drug resistance testing ∇. J. Clin. Microbiol. 2009;47(4):1107–1118. doi: 10.1128/JCM.02255-08. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous