p39-associated Cdk5 activity regulates dendritic morphogenesis
- PMID: 33127972
- PMCID: PMC7603351
- DOI: 10.1038/s41598-020-75264-6
p39-associated Cdk5 activity regulates dendritic morphogenesis
Abstract
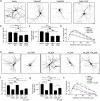
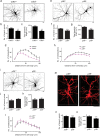
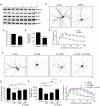
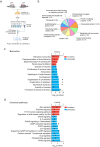
Dendrites, branched structures extending from neuronal cell soma, are specialized for processing information from other neurons. The morphogenesis of dendritic structures is spatiotemporally regulated by well-orchestrated signaling cascades. Dysregulation of these processes impacts the wiring of neuronal circuit and efficacy of neurotransmission, which contribute to the pathogeneses of neurological disorders. While Cdk5 (cyclin-dependent kinase 5) plays a critical role in neuronal dendritic development, its underlying molecular control is not fully understood. In this study, we show that p39, one of the two neuronal Cdk5 activators, is a key regulator of dendritic morphogenesis. Pyramidal neurons deficient in p39 exhibit aberrant dendritic morphology characterized by shorter length and reduced arborization, which is comparable to dendrites in Cdk5-deficient neurons. RNA sequencing analysis shows that the adaptor protein, WDFY1 (WD repeat and FYVE domain-containing 1), acts downstream of Cdk5/p39 to regulate dendritic morphogenesis. While WDFY1 is elevated in p39-deficient neurons, suppressing its expression rescues the impaired dendritic arborization. Further phosphoproteomic analysis suggests that Cdk5/p39 mediates dendritic morphogenesis by modulating various downstream signaling pathways, including PI3K/Akt-, cAMP-, or small GTPase-mediated signaling transduction pathways, thereby regulating cytoskeletal organization, protein synthesis, and protein trafficking.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

