Retrograde signaling by a mtDNA-encoded non-coding RNA preserves mitochondrial bioenergetics
- PMID: 33127975
- PMCID: PMC7603330
- DOI: 10.1038/s42003-020-01322-4
Retrograde signaling by a mtDNA-encoded non-coding RNA preserves mitochondrial bioenergetics
Abstract
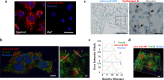
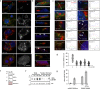
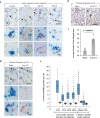
Alveolar epithelial type II (AETII) cells are important for lung epithelium maintenance and function. We demonstrate that AETII cells from mouse lungs exposed to cigarette smoke (CS) increase the levels of the mitochondria-encoded non-coding RNA, mito-RNA-805, generated by the control region of the mitochondrial genome. The protective effects of mito-ncR-805 are associated with positive regulation of mitochondrial energy metabolism, and respiration. Levels of mito-ncR-805 do not relate to steady-state transcription or replication of the mitochondrial genome. Instead, CS-exposure causes the redistribution of mito-ncR-805 from mitochondria to the nucleus, which correlated with the increased expression of nuclear-encoded genes involved in mitochondrial function. These studies reveal an unrecognized mitochondria stress associated retrograde signaling, and put forward the idea that mito-ncRNA-805 represents a subtype of small non coding RNAs that are regulated in a tissue- or cell-type specific manner to protect cells under physiological stress.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Mitochondrial ncRNA LDL-805 declines in alveolar epithelial type 2 cells of chronic obstructive pulmonary disease patients.bioRxiv [Preprint]. 2024 Jan 15:2024.01.14.575591. doi: 10.1101/2024.01.14.575591. bioRxiv. 2024. PMID: 38293158 Free PMC article. Preprint.
-
A Synthetic Small RNA Homologous to the D-Loop Transcript of mtDNA Enhances Mitochondrial Bioenergetics.Front Physiol. 2022 Apr 6;13:772313. doi: 10.3389/fphys.2022.772313. eCollection 2022. Front Physiol. 2022. PMID: 35464086 Free PMC article.
-
Mitochondrial 8-oxoguanine DNA glycosylase mitigates alveolar epithelial cell PINK1 deficiency, mitochondrial DNA damage, apoptosis, and lung fibrosis.Am J Physiol Lung Cell Mol Physiol. 2020 May 1;318(5):L1084-L1096. doi: 10.1152/ajplung.00069.2019. Epub 2020 Mar 25. Am J Physiol Lung Cell Mol Physiol. 2020. PMID: 32209025 Free PMC article.
-
Decoding mitochondrial-nuclear (epi)genome interactions: the emerging role of ncRNAs.Epigenomics. 2023 Nov;15(21):1121-1136. doi: 10.2217/epi-2023-0322. Epub 2023 Nov 21. Epigenomics. 2023. PMID: 38031736 Review.
-
Maintenance and Expression of Mammalian Mitochondrial DNA.Annu Rev Biochem. 2016 Jun 2;85:133-60. doi: 10.1146/annurev-biochem-060815-014402. Epub 2016 Mar 24. Annu Rev Biochem. 2016. PMID: 27023847 Review.
Cited by
-
Editorial: Role of mitochondria-associated non-coding RNAs in intracellular communication.Front Physiol. 2022 Aug 22;13:980674. doi: 10.3389/fphys.2022.980674. eCollection 2022. Front Physiol. 2022. PMID: 36072845 Free PMC article. No abstract available.
-
Guidelines for mitochondrial RNA analysis.Mol Ther Nucleic Acids. 2024 Jun 26;35(3):102262. doi: 10.1016/j.omtn.2024.102262. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39091381 Free PMC article. Review.
-
MicroRNAs as Factors in Bidirectional Crosstalk Between Mitochondria and the Nucleus During Cellular Senescence.Front Physiol. 2021 Sep 9;12:734976. doi: 10.3389/fphys.2021.734976. eCollection 2021. Front Physiol. 2021. PMID: 34566699 Free PMC article. Review.
-
Mitochondria - the CEO of the cell.J Cell Sci. 2025 May 1;138(9):jcs263403. doi: 10.1242/jcs.263403. Epub 2025 May 1. J Cell Sci. 2025. PMID: 40310473 Review.
-
Redox signaling-mediated S-glutathionylation of protein disulfide isomerase A1 initiates intrinsic apoptosis and contributes to accelerated aging.Redox Biol. 2025 May 27;85:103680. doi: 10.1016/j.redox.2025.103680. Online ahead of print. Redox Biol. 2025. PMID: 40472774 Free PMC article.
References
-
- National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General Reports of the Surgeon General (Centers for Disease Control and Prevention, Atlanta, GA, 2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

