System-level analyses of keystone genes required for mammalian tooth development
- PMID: 33128445
- PMCID: PMC7894285
- DOI: 10.1002/jez.b.23009
System-level analyses of keystone genes required for mammalian tooth development
Abstract
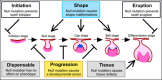
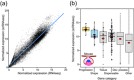
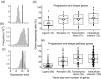
When a null mutation of a gene causes a complete developmental arrest, the gene is typically considered essential for life. Yet, in most cases, null mutations have more subtle effects on the phenotype. Here we used the phenotypic severity of mutations as a tool to examine system-level dynamics of gene expression. We classify genes required for the normal development of the mouse molar into different categories that range from essential to subtle modification of the phenotype. Collectively, we call these the developmental keystone genes. Transcriptome profiling using microarray and RNAseq analyses of patterning stage mouse molars show highly elevated expression levels for genes essential for the progression of tooth development, a result reminiscent of essential genes in single-cell organisms. Elevated expression levels of progression genes were also detected in developing rat molars, suggesting evolutionary conservation of this system-level dynamics. Single-cell RNAseq analyses of developing mouse molars reveal that even though the size of the expression domain, measured in the number of cells, is the main driver of organ-level expression, progression genes show high cell-level transcript abundances. Progression genes are also upregulated within their pathways, which themselves are highly expressed. In contrast, a high proportion of the genes required for normal tooth patterning are secreted ligands that are expressed in fewer cells than their receptors and intracellular components. Overall, even though expression patterns of individual genes can be highly different, conserved system-level principles of gene expression can be detected using phenotypically defined gene categories.
Keywords: essential genes; keystone genes; single-cell RNAseq; tooth development; transcript abundance; transcriptomes.
© 2020 The Authors. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution Published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there is no conflict of interests.
Figures



Similar articles
-
How do genes make teeth to order through development?J Exp Zool B Mol Dev Evol. 2006 May 15;306(3):177-82. doi: 10.1002/jez.b.21104. J Exp Zool B Mol Dev Evol. 2006. PMID: 16615103 Review.
-
Bank vole genomics links determinate and indeterminate growth of teeth.BMC Genomics. 2024 Oct 30;25(1):1000. doi: 10.1186/s12864-024-10901-2. BMC Genomics. 2024. PMID: 39472825 Free PMC article.
-
Essential roles of G9a in cell proliferation and differentiation during tooth development.Exp Cell Res. 2017 Aug 15;357(2):202-210. doi: 10.1016/j.yexcr.2017.05.016. Epub 2017 May 17. Exp Cell Res. 2017. PMID: 28527696
-
Phylogenetic memory of developing mammalian dentition.J Exp Zool B Mol Dev Evol. 2006 May 15;306(3):234-50. doi: 10.1002/jez.b.21093. J Exp Zool B Mol Dev Evol. 2006. PMID: 16463376 Review.
-
Insight from Frogs: Sonic Hedgehog Gene Expression and a Re-evaluation of the Vertebrate Odontogenic Band.Anat Rec (Hoboken). 2016 Aug;299(8):1099-109. doi: 10.1002/ar.23378. Epub 2016 Jun 20. Anat Rec (Hoboken). 2016. PMID: 27262165
Cited by
-
Unraveling the hidden complexity: Exploring dental tissues through single-cell transcriptional profiling.Regen Ther. 2024 Apr 2;27:218-229. doi: 10.1016/j.reth.2024.03.023. eCollection 2024 Dec. Regen Ther. 2024. PMID: 38596822 Free PMC article. Review.
-
An epithelial signalling centre in sharks supports homology of tooth morphogenesis in vertebrates.Elife. 2022 May 10;11:e73173. doi: 10.7554/eLife.73173. Elife. 2022. PMID: 35536602 Free PMC article.
-
The Secretome of the Inductive Tooth Germ Exhibits Signals Required for Tooth Development.Bioengineering (Basel). 2025 Jan 21;12(2):96. doi: 10.3390/bioengineering12020096. Bioengineering (Basel). 2025. PMID: 40001617 Free PMC article.
-
Mapping molar shapes on signaling pathways.PLoS Comput Biol. 2020 Dec 14;16(12):e1008436. doi: 10.1371/journal.pcbi.1008436. eCollection 2020 Dec. PLoS Comput Biol. 2020. PMID: 33315865 Free PMC article.
-
Distinct BMP-Smad signaling outputs confer diverse functions in dental mesenchyme.Development. 2025 Jun 15;152(12):dev204563. doi: 10.1242/dev.204563. Epub 2025 Jun 19. Development. 2025. PMID: 40446216
References
-
- Abouheif, E. , & Wray, G. (2002). Evolution of the gene network underlying wing polyphenism in ants. Science, 297, 249–252. - PubMed
-
- Amsterdam, A. , Nissen, R. M. , Sun, Z. , Swindell, E. C. , Farrington, S. , & Hopkins, N. (2004). Identification of 315 genes essential for early zebrafish development. Proceedings of the National Academy of Sciences of the United States of America, 101, 12792–12797. 10.1073/pnas.0403929101 - DOI - PMC - PubMed
-
- Andrews, S. , Krueger, F. , Segonds‐Pichon, A. , Biggins, L. , Krueger, C. , & Wingett, S. (2012). FastQC. Babraham: Babraham Institute.
-
- Ashburner, M. , Ball, C. A. , Blake, J. A. , Botstein, D. , Butler, H. , Cherry, J. M. , Davis, A. P. , Dolinski, K. , Dwight, S. S. , Eppig, J. T. , Harris, M. A. , Hill, D. P. , Issel‐Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J. C. , Richardson, J. E. , Ringwald, M. , Rubin, G. M. , & Sherlock, G. (2000). Gene Ontology: Tool for the unification of biology. Nature Genetics, 25, 25–29. 10.1038/75556 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

