Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes
- PMID: 33128788
- PMCID: PMC11979674
- DOI: 10.1111/myc.13204
Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes
Abstract
Background: The pathogen Candida auris is rapidly gaining clinical importance because of its resistance to antifungal treatments and its persistence in hospital environments. Early and accurate diagnosis of C. auris infections is crucial, and however, the fungus has often been misidentified by commercial systems.
Objectives: To develop conventional and real-time PCR methods for accurate and rapid identification of C. auris and its discrimination from closely related species by exploiting the uniqueness of certain glycosylphosphatidylinositol (GPI)-modified protein-encoding genes.
Methods: Species-specific primers for two unique putative GPI protein-encoding genes per species were designed for C. auris, C. haemulonii, C. pseudohaemulonii, C. duobushaemulonii, C. lusitaniae and C. albicans. Primers were blind tested for their specificity and efficiency in conventional and real-time multiplex PCR set-up.
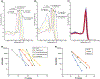
Results: All primers combinations showed excellent species specificity. In multiplex mode, correct identification was aided by different-sized amplicons for each species. Efficiency of the C. auris primers was validated using a panel of 155 C. auris isolates, including all known genetically diverse clades. In real-time multiplex PCR, different melting points of the amplicons allowed the distinction of C. auris from four related species. C. auris limit of detection was 5 CFU/reaction with a threshold value of 32. The method was also able to detect C. auris in spiked blood and serum.
Conclusions: PCR identification based on unique GPI protein-encoding genes allows for accurate and rapid species identification of C. auris and related species without need for expensive equipment when applied in conventional PCR set-up.
Keywords: Candida; Candida auris; C. haemulonii complex; PCR identification; candidosis; yeasts.
© 2020 Wiley-VCH GmbH.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures

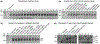
References
-
- Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009; 53: 41–44. - PubMed
-
- Kim MN, Shin JH, Sung H, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009; 48: e57–e61. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

