Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction
- PMID: 33128895
- PMCID: PMC7577733
- DOI: 10.1016/j.stem.2020.10.005
Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction
Abstract
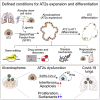
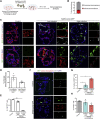
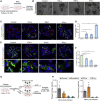
Coronavirus infection causes diffuse alveolar damage leading to acute respiratory distress syndrome. The absence of ex vivo models of human alveolar epithelium is hindering an understanding of coronavirus disease 2019 (COVID-19) pathogenesis. Here, we report a feeder-free, scalable, chemically defined, and modular alveolosphere culture system for the propagation and differentiation of human alveolar type 2 cells/pneumocytes derived from primary lung tissue. Cultured pneumocytes express the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor angiotensin-converting enzyme receptor type-2 (ACE2) and can be infected with virus. Transcriptome and histological analysis of infected alveolospheres mirror features of COVID-19 lungs, including emergence of interferon (IFN)-mediated inflammatory responses, loss of surfactant proteins, and apoptosis. Treatment of alveolospheres with IFNs recapitulates features of virus infection, including cell death. In contrast, alveolospheres pretreated with low-dose IFNs show a reduction in viral replication, suggesting the prophylactic effectiveness of IFNs against SARS-CoV-2. Human stem cell-based alveolospheres, thus, provide novel insights into COVID-19 pathogenesis and can serve as a model for understanding human respiratory diseases.
Keywords: ACE2; ARDS; SARS-CoV-2; cytokine storm; interferons; organoids; pneumocytes; protease; respiratory cells; surfactants.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A patent application (PCT/US20/53158) related to this work has been filed. H.K. and P.R.T. are listed as co-inventors on this application. P.R.T. serves as a consultant for Cellarity and Surrozen.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL153375/HL/NHLBI NIH HHS/United States
- R21 GM131279/GM/NIGMS NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R00 HL127181/HL/NHLBI NIH HHS/United States
- F30 HL143911/HL/NHLBI NIH HHS/United States
- U01 AI149644/AI/NIAID NIH HHS/United States
- R01 HL146557/HL/NHLBI NIH HHS/United States
- P01 HL108808/HL/NHLBI NIH HHS/United States
- P30 DK065988/DK/NIDDK NIH HHS/United States
- UC6 AI058607/AI/NIAID NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
- T32 GM007171/GM/NIGMS NIH HHS/United States
- R01 AI132178/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

