OMA1-An integral membrane protease?
- PMID: 33130089
- PMCID: PMC7770061
- DOI: 10.1016/j.bbapap.2020.140558
OMA1-An integral membrane protease?
Abstract
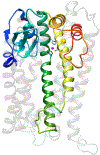
OMA1 is a mitochondrial protease. Among its substrates are DELE1, a signaling peptide, which can elicit the integrated stress response, as well as the membrane-shaping dynamin-related GTPase OPA1, which can drive mitochondrial outer membrane permeabilization. OMA1 is dormant under physiological conditions but rapidly activated upon mitochondrial stress, such as loss of membrane potential or excessive reactive oxygen species. Accordingly, OMA1 was found to be activated in a number of disease conditions, including cancer and neurodegeneration. OMA1 has a predicted transmembrane domain and is believed to be tethered to the mitochondrial inner membrane. Yet, its structure has not been resolved and its context-dependent regulation remains obscure. Here, I review the literature with focus on OMA1's biochemistry. I provide a good homology model of OMA1's active site with a root-mean-square deviation of 0.9 Å and a DALI Z-score of 19.8. And I build a case for OMA1 actually being an integral membrane protease based on OMA1's role in the generation of small signaling peptides, its functional overlap with PARL, and OMA1's homology with ZMPSTE24. The refined understanding of this important enzyme can help with the design of tool compounds and development of chemical probes in the future.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
Dr. Marcel V. Alavi is shareholder of 712 North Inc., a California-based pharmaceutical company.
Figures

References
-
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D, An integrated stress response regulates amino acid metabolism and resistance to oxidative stress, Mol Cell 11 (2003) 619–633. - PubMed
-
- Kaser M, Kambacheld M, Kisters-Woike B, Langer T, Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease, J Biol Chem 278 (2003) 46414–46423. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

