Decoding mixed messages in the developing cortex: translational regulation of neural progenitor fate
- PMID: 33130411
- PMCID: PMC8058166
- DOI: 10.1016/j.conb.2020.10.001
Decoding mixed messages in the developing cortex: translational regulation of neural progenitor fate
Abstract
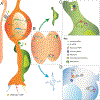
Regulation of stem cell fate decisions is elemental to faithful development, homeostasis, and organismal fitness. Emerging data demonstrate pluripotent stem cells exhibit a vast transcriptional landscape, which is refined as cells differentiate. In the developing neocortex, transcriptional priming of neural progenitors, coupled with post-transcriptional control, is critical for defining cell fates of projection neurons. In particular, radial glial progenitors exhibit dynamic post-transcriptional regulation, including subcellular mRNA localization, RNA decay, and translation. These processes involve both cis-regulatory and trans-regulatory factors, many of which are implicated in neurodevelopmental disease. This review highlights emerging post-transcriptional mechanisms which govern cortical development, with a particular focus on translational control of neuronal fates, including those relevant for disease.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
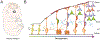
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

