Ubiquitin signaling in cell cycle control and tumorigenesis
- PMID: 33130827
- PMCID: PMC7862229
- DOI: 10.1038/s41418-020-00648-0
Ubiquitin signaling in cell cycle control and tumorigenesis
Abstract
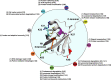
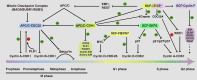
Cell cycle progression is a tightly regulated process by which DNA replicates and cell reproduces. The major driving force underlying cell cycle progression is the sequential activation of cyclin-dependent kinases (CDKs), which is achieved in part by the ubiquitin-mediated proteolysis of their cyclin partners and kinase inhibitors (CKIs). In eukaryotic cells, two families of E3 ubiquitin ligases, anaphase-promoting complex/cyclosome and Skp1-Cul1-F-box protein complex, are responsible for ubiquitination and proteasomal degradation of many of these CDK regulators, ensuring cell cycle progresses in a timely and precisely regulated manner. In the past couple of decades, accumulating evidence have demonstrated that the dysregulated cell cycle transition caused by inefficient proteolytic control leads to uncontrolled cell proliferation and finally results in tumorigenesis. Based upon this notion, targeting the E3 ubiquitin ligases involved in cell cycle regulation is expected to provide novel therapeutic strategies for cancer treatment. Thus, a better understanding of the diversity and complexity of ubiquitin signaling in cell cycle regulation will shed new light on the precise control of the cell cycle progression and guide anticancer drug development.
Conflict of interest statement
WW is a co-founder and consultant for the ReKindle Therapeutics. Other authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

