Functional analysis of low-grade glioma genetic variants predicts key target genes and transcription factors
- PMID: 33130899
- PMCID: PMC8041333
- DOI: 10.1093/neuonc/noaa248
Functional analysis of low-grade glioma genetic variants predicts key target genes and transcription factors
Abstract
Background: Large-scale genome-wide association studies (GWAS) have implicated thousands of germline genetic variants in modulating individuals' risk to various diseases, including cancer. At least 25 risk loci have been identified for low-grade gliomas (LGGs), but their molecular functions remain largely unknown.
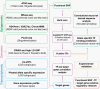
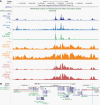
Methods: We hypothesized that GWAS loci contain causal single nucleotide polymorphisms (SNPs) that reside in accessible open chromatin regions and modulate the expression of target genes by perturbing the binding affinity of transcription factors (TFs). We performed an integrative analysis of genomic and epigenomic data from The Cancer Genome Atlas and other public repositories to identify candidate causal SNPs within linkage disequilibrium blocks of LGG GWAS loci. We assessed their potential regulatory role via in silico TF binding sequence perturbations, convolutional neural network trained on TF binding data, and simulated annealing-based interpretation methods.
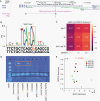
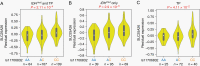
Results: We built an interactive website (http://education.knoweng.org/alg3/) summarizing the functional footprinting of 280 variants in 25 LGG GWAS regions, providing rich information for further computational and experimental scrutiny. We identified as case studies PHLDB1 and SLC25A26 as candidate target genes of rs12803321 and rs11706832, respectively, and predicted the GWAS variant rs648044 to be the causal SNP modulating ZBTB16, a known tumor suppressor in multiple cancers. We showed that rs648044 likely perturbed the binding affinity of the TF MAFF, as supported by RNA interference and in vitro MAFF binding experiments.
Conclusions: The identified candidate (causal SNP, target gene, TF) triplets and the accompanying resource will help accelerate our understanding of the molecular mechanisms underlying genetic risk factors for gliomas.
Keywords: GWAS; functional genomics; genetic variants; low-grade glioma.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. - PubMed

