Characterization of Organoid Cultures to Study the Effects of Pregnancy Hormones on the Epigenome and Transcriptional Output of Mammary Epithelial Cells
- PMID: 33131024
- PMCID: PMC7960614
- DOI: 10.1007/s10911-020-09465-0
Characterization of Organoid Cultures to Study the Effects of Pregnancy Hormones on the Epigenome and Transcriptional Output of Mammary Epithelial Cells
Abstract
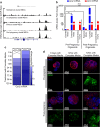
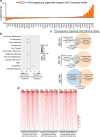
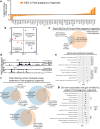
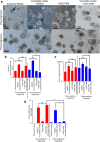
The use of mouse derived mammary organoids can provide a unique strategy to study mammary gland development across a normal life cycle, as well as offering insights into how malignancies form and progress. Substantial cellular and epigenomic changes are triggered in response to pregnancy hormones, a reaction that engages molecular and cellular changes that transform the mammary epithelial cells into "milk producing machines". Such epigenomic alterations remain stable in post-involution mammary epithelial cells and control the reactivation of gene transcription in response to re-exposure to pregnancy hormones. Thus, a system that tightly controls exposure to pregnancy hormones, epigenomic alterations, and activation of transcription will allow for a better understanding of such molecular switches. Here, we describe the characterization of ex vivo cultures to mimic the response of mammary organoid cultures to pregnancy hormones and to understand gene regulation and epigenomic reprogramming on consecutive hormone exposure. Our findings suggest that this system yields similar epigenetic modifications to those reported in vivo, thus representing a suitable model to closely track epigenomic rearrangement and define unknown players of pregnancy-induced development.
Keywords: Epigenomics; Mammary organoids; Pregnancy-induced development.
Conflict of interest statement
The authors have no competing interests to disclose.
Figures
Similar articles
-
Alveolar progenitor cells develop in mouse mammary glands independent of pregnancy and lactation.J Cell Physiol. 2007 Sep;212(3):729-36. doi: 10.1002/jcp.21071. J Cell Physiol. 2007. PMID: 17443685
-
Estradiol, progesterone and prolactin modulate mammary gland morphogenesis in adult female plains vizcacha (Lagostomus maximus).J Mol Histol. 2013 Jun;44(3):299-310. doi: 10.1007/s10735-012-9477-0. Epub 2013 Mar 26. J Mol Histol. 2013. PMID: 23529757
-
Role of ovarian secretions in mammary gland development and function in ruminants.Animal. 2014 Jan;8(1):72-85. doi: 10.1017/S1751731113001638. Epub 2013 Oct 8. Animal. 2014. PMID: 24103527 Review.
-
Hormonal regulation of miRNA during mammary gland development.Biol Open. 2024 Jun 15;13(6):bio060308. doi: 10.1242/bio.060308. Epub 2024 Jun 10. Biol Open. 2024. PMID: 38712984 Free PMC article.
-
Review: Mammary gland development in swine: embryo to early lactation.Animal. 2019 Jul;13(S1):s11-s19. doi: 10.1017/S1751731119000521. Animal. 2019. PMID: 31280748 Review.
Cited by
-
Macrophages maintain mammary stem cell activity and mammary homeostasis via TNF-α-PI3K-Cdk1/Cyclin B1 axis.NPJ Regen Med. 2023 May 2;8(1):23. doi: 10.1038/s41536-023-00296-1. NPJ Regen Med. 2023. PMID: 37130846 Free PMC article.
-
Unraveling the Breast: Advances in Mammary Biology and Cancer Methods.J Mammary Gland Biol Neoplasia. 2020 Dec;25(4):233-236. doi: 10.1007/s10911-020-09476-x. Epub 2021 Jan 21. J Mammary Gland Biol Neoplasia. 2020. PMID: 33479879 Free PMC article.
-
Clinical applications of 3D normal and breast cancer organoids: A review of concepts and methods.Exp Biol Med (Maywood). 2022 Dec;247(24):2176-2183. doi: 10.1177/15353702221131877. Epub 2022 Nov 19. Exp Biol Med (Maywood). 2022. PMID: 36408534 Free PMC article. Review.
-
The molecular basis of mammary gland development and epithelial differentiation.Semin Cell Dev Biol. 2021 Jun;114:93-112. doi: 10.1016/j.semcdb.2020.09.014. Epub 2020 Oct 17. Semin Cell Dev Biol. 2021. PMID: 33082117 Free PMC article. Review.
-
Gestational Breast Cancer - a Review of Outcomes, Pathophysiology, and Model Systems.J Mammary Gland Biol Neoplasia. 2023 Jul 14;28(1):16. doi: 10.1007/s10911-023-09546-w. J Mammary Gland Biol Neoplasia. 2023. PMID: 37450228 Free PMC article. Review.
References
-
- Ewald AJ. Isolation of mouse mammary organoids for long-term time-lapse imaging. Cold Spring Harb Protoc. 2013;2013(2):130–133. - PubMed
-
- Lo AT, Mori H, Mott J, Bissell MJ. Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J Mammary Gland Biol Neoplasia. 2012;17(2):103–110. - PubMed
-
- Reginato MJ, Muthuswamy SK. Illuminating the center: mechanisms regulating lumen formation and maintenance in mammary morphogenesis. J Mammary Gland Biol Neoplasia. 2006;11(3–4):205–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

