A behavioral model for mapping the genetic architecture of gut-microbiota networks
- PMID: 33131416
- PMCID: PMC8381822
- DOI: 10.1080/19490976.2020.1820847
A behavioral model for mapping the genetic architecture of gut-microbiota networks
Abstract
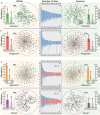
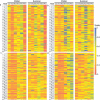
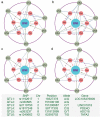
The gut microbiota may play an important role in affecting human health. To explore the genetic mechanisms underlying microbiota-host relationships, many genome-wide association studies have begun to identify host genes that shape the microbial composition of the gut. It is becoming increasingly clear that the gut microbiota impacts host processes not only through the action of individual microbes but also their interaction networks. However, a systematic characterization of microbial interactions that occur in densely packed aggregates of the gut bacteria has proven to be extremely difficult. We develop a computational rule of thumb for addressing this issue by integrating ecological behavioral theory and genetic mapping theory. We introduce behavioral ecology theory to derive mathematical descriptors of how each microbe interacts with every other microbe through a web of cooperation and competition. We estimate the emergent properties of gut-microbiota networks reconstructed from these descriptors and map host-driven mutualism, antagonism, aggression, and altruism QTLs. We further integrate path analysis and mapping theory to detect and visualize how host genetic variants affect human diseases by perturbing the internal workings of the gut microbiota. As the proof of concept, we apply our model to analyze a published dataset of the gut microbiota, showing its usefulness and potential to gain new insight into how microbes are organized in human guts. The new model provides an analytical tool for revealing the "endophenotype" role of microbial networks in linking genotype to end-point phenotypes.
Keywords: The gut microbiota; competition; cooperation; network science; qtl; rule of thumb.
Figures

Similar articles
-
Disentangling leaf-microbiome interactions in Arabidopsis thaliana by network mapping.Front Plant Sci. 2022 Oct 6;13:996121. doi: 10.3389/fpls.2022.996121. eCollection 2022. Front Plant Sci. 2022. PMID: 36275601 Free PMC article.
-
Network mapping of root-microbe interactions in Arabidopsis thaliana.NPJ Biofilms Microbiomes. 2021 Sep 7;7(1):72. doi: 10.1038/s41522-021-00241-4. NPJ Biofilms Microbiomes. 2021. PMID: 34493731 Free PMC article.
-
Universal gut microbial relationships in the gut microbiome of wild baboons.Elife. 2023 May 9;12:e83152. doi: 10.7554/eLife.83152. Elife. 2023. PMID: 37158607 Free PMC article.
-
Understanding Competition and Cooperation within the Mammalian Gut Microbiome.Curr Biol. 2019 Jun 3;29(11):R538-R544. doi: 10.1016/j.cub.2019.04.017. Curr Biol. 2019. PMID: 31163167 Free PMC article. Review.
-
The opportunistic nature of gut commensal microbiota.Crit Rev Microbiol. 2023 Nov;49(6):739-763. doi: 10.1080/1040841X.2022.2133987. Epub 2022 Oct 18. Crit Rev Microbiol. 2023. PMID: 36256871 Review.
Cited by
-
Disentangling leaf-microbiome interactions in Arabidopsis thaliana by network mapping.Front Plant Sci. 2022 Oct 6;13:996121. doi: 10.3389/fpls.2022.996121. eCollection 2022. Front Plant Sci. 2022. PMID: 36275601 Free PMC article.
-
Integrated multi-omics of the gastrointestinal microbiome and ruminant host reveals metabolic adaptation underlying early life development.Microbiome. 2022 Dec 12;10(1):222. doi: 10.1186/s40168-022-01396-8. Microbiome. 2022. PMID: 36503572 Free PMC article.
-
Microbiome-based classification models for fresh produce safety and quality evaluation.Microbiol Spectr. 2024 Apr 2;12(4):e0344823. doi: 10.1128/spectrum.03448-23. Epub 2024 Mar 6. Microbiol Spectr. 2024. PMID: 38445872 Free PMC article.
-
Modeling spatial interaction networks of the gut microbiota.Gut Microbes. 2022 Jan-Dec;14(1):2106103. doi: 10.1080/19490976.2022.2106103. Gut Microbes. 2022. PMID: 35921525 Free PMC article.
-
Hypernetwork modeling and topology of high-order interactions for complex systems.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2412220121. doi: 10.1073/pnas.2412220121. Epub 2024 Sep 24. Proc Natl Acad Sci U S A. 2024. PMID: 39316048 Free PMC article.
References
-
- Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–1412. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
