The Unfolded Protein Response and Autophagy as Drug Targets in Neuropsychiatric Disorders
- PMID: 33132844
- PMCID: PMC7550790
- DOI: 10.3389/fncel.2020.554548
The Unfolded Protein Response and Autophagy as Drug Targets in Neuropsychiatric Disorders
Abstract
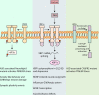
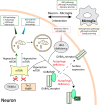
Neurons are polarized in structure with a cytoplasmic compartment extending into dendrites and a long axon that terminates at the synapse. The high level of compartmentalization imposes specific challenges for protein quality control in neurons making them vulnerable to disturbances that may lead to neurological dysfunctions including neuropsychiatric diseases. Synapse and dendrites undergo structural modulations regulated by neuronal activity involve key proteins requiring strict control of their turnover rates and degradation pathways. Recent advances in the study of the unfolded protein response (UPR) and autophagy processes have brought novel insights into the specific roles of these processes in neuronal physiology and synaptic signaling. In this review, we highlight recent data and concepts about UPR and autophagy in neuropsychiatric disorders and synaptic plasticity including a brief outline of possible therapeutic approaches to influence UPR and autophagy signaling in these diseases.
Keywords: UPR; autophagy; drug; neuropsychiatric disease; synapse.
Copyright © 2020 Srinivasan, Korhonen and Lindholm.
Figures
Similar articles
-
Interplay between unfolded protein response and autophagy promotes tumor drug resistance.Oncol Lett. 2015 Oct;10(4):1959-1969. doi: 10.3892/ol.2015.3508. Epub 2015 Jul 17. Oncol Lett. 2015. PMID: 26622781 Free PMC article.
-
Synapse-to-Nucleus Signaling in Neurodegenerative and Neuropsychiatric Disorders.Biol Psychiatry. 2019 Jul 15;86(2):87-96. doi: 10.1016/j.biopsych.2019.01.006. Epub 2019 Jan 17. Biol Psychiatry. 2019. PMID: 30846302 Review.
-
The broad spectrum of signaling pathways regulated by unfolded protein response in neuronal homeostasis.Neurochem Int. 2018 Oct;119:26-34. doi: 10.1016/j.neuint.2017.06.012. Epub 2017 Jun 28. Neurochem Int. 2018. PMID: 28668471 Review.
-
Emerging Concepts and Functions of Autophagy as a Regulator of Synaptic Components and Plasticity.Cells. 2019 Jan 9;8(1):34. doi: 10.3390/cells8010034. Cells. 2019. PMID: 30634508 Free PMC article. Review.
-
Recent Insights into the Role of Unfolded Protein Response in ER Stress in Health and Disease.Front Cell Dev Biol. 2017 May 10;5:48. doi: 10.3389/fcell.2017.00048. eCollection 2017. Front Cell Dev Biol. 2017. PMID: 28540288 Free PMC article. Review.
Cited by
-
Inflammatory signaling mechanisms in bipolar disorder.J Biomed Sci. 2021 Jun 11;28(1):45. doi: 10.1186/s12929-021-00742-6. J Biomed Sci. 2021. PMID: 34112182 Free PMC article. Review.
-
Neuroinflammatory Response and Redox-regulation Activity of Hyperoside in Manganese-induced Neurotoxicity Model of Wistar Rats.Curr Aging Sci. 2024;17(3):220-236. doi: 10.2174/0118746098277166231204103616. Curr Aging Sci. 2024. PMID: 38500281
-
A dominant negative Kcnd3 F227del mutation in mice causes spinocerebellar ataxia type 22 (SCA22) by impairing ER and Golgi functioning.J Pathol. 2025 Jan;265(1):57-68. doi: 10.1002/path.6368. Epub 2024 Nov 19. J Pathol. 2025. PMID: 39562497 Free PMC article.
-
Recent Developments in Protein Lactylation in PTSD and CVD: Novel Strategies and Targets.BioTech (Basel). 2023 May 15;12(2):38. doi: 10.3390/biotech12020038. BioTech (Basel). 2023. PMID: 37218755 Free PMC article. Review.
-
Sphingolipids as Modulators of SARS-CoV-2 Infection.Front Cell Dev Biol. 2021 Jun 17;9:689854. doi: 10.3389/fcell.2021.689854. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34222257 Free PMC article. Review.
References
-
- Abelaira H. M., Reus G. Z., Ignacio Z. M., Dos Santos M. A., de Moura A. B., Matos D., et al. . (2017). Effects of ketamine administration on mTOR and reticulum stress signaling pathways in the brain after the infusion of rapamycin into prefrontal cortex. J. Psychiatr. Res. 87, 81–87. 10.1016/j.jpsychires.2016.12.002 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

