circFAT1(e2) Promotes Papillary Thyroid Cancer Proliferation, Migration, and Invasion via the miRNA-873/ZEB1 Axis
- PMID: 33133224
- PMCID: PMC7593750
- DOI: 10.1155/2020/1459368
circFAT1(e2) Promotes Papillary Thyroid Cancer Proliferation, Migration, and Invasion via the miRNA-873/ZEB1 Axis
Retraction in
-
Retracted: circFAT1(e2) Promotes Papillary Thyroid Cancer Proliferation, Migration, and Invasion via the miRNA-873/ZEB1 Axis.Comput Math Methods Med. 2023 Oct 18;2023:9820923. doi: 10.1155/2023/9820923. eCollection 2023. Comput Math Methods Med. 2023. PMID: 37885876 Free PMC article.
Abstract
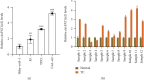
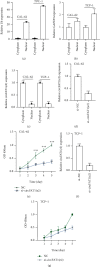
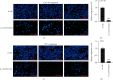
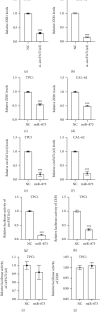
Circular RNAs (circRNAs) play an extremely important regulatory role in the occurrence and development of various malignant tumors including papillary thyroid cancer (PTC). circFAT1(e2) is a new type of circRNA derived from exon 2 of the FAT1 gene, which is distributed in the cytoplasm and nucleus of PTC cells. However, so far, the role of circFAT1(e2) in PTC is still unclear. In this study, circFAT1(e2) was found to be highly expressed in PTC cell lines and tissues. circFAT1(e2) knockdown suppressed PTC cell growth, migration, and invasion. Also, circFAT1(e2) acted as a sponge for potential microRNAs (miRNAs) to modulate cancer progression. A potential miRNA target was discovered to be miR-873 which was targeted by circFAT1(e2) in PTC. The dual-luciferase assay conducted later also confirmed that there was indeed a direct interaction between circFAT1(e2) and miR-873. This study also confirmed that circFAT1(e2) inhibited the miR-873 expression and thus promoted the ZEB1 expression, thus affecting the proliferation, metastasis, and invasion of PTC cells. In conclusion, the results of this study indicated that circFAT1(e2) played a carcinogenic role by targeting the miR-873/ZEB1 axis to promote PTC invasion and metastasis, which might become a potential novel target for therapy of PTC.
Copyright © 2020 Jiazhe Liu et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Circular RNA circFAT1(e2) Promotes Colorectal Cancer Tumorigenesis via the miR-30e-5p/ITGA6 Axis.Comput Math Methods Med. 2021 Jun 26;2021:9980459. doi: 10.1155/2021/9980459. eCollection 2021. Comput Math Methods Med. 2021. Retraction in: Comput Math Methods Med. 2023 Sep 27;2023:9763752. doi: 10.1155/2023/9763752. PMID: 34257702 Free PMC article. Retracted.
-
Long noncoding RNA SNHG22 increases ZEB1 expression via competitive binding with microRNA-429 to promote the malignant development of papillary thyroid cancer.Cell Cycle. 2020 May;19(10):1186-1199. doi: 10.1080/15384101.2020.1749466. Epub 2020 Apr 19. Cell Cycle. 2020. Retraction in: Cell Cycle. 2021 Dec;20(24):2667. doi: 10.1080/15384101.2021.1971879. PMID: 32306838 Free PMC article. Retracted.
-
Exosomal circular RNA hsa_circ_007293 promotes proliferation, migration, invasion, and epithelial-mesenchymal transition of papillary thyroid carcinoma cells through regulation of the microRNA-653-5p/paired box 6 axis.Bioengineered. 2021 Dec;12(2):10136-10149. doi: 10.1080/21655979.2021.2000745. Bioengineered. 2021. PMID: 34866540 Free PMC article.
-
The Emerging Roles of circRNAs in Papillary Thyroid Carcinoma: Molecular Mechanisms and Biomarker Potential.Protein Pept Lett. 2023;30(9):709-718. doi: 10.2174/0929866530666230804104057. Protein Pept Lett. 2023. PMID: 37537939 Review.
-
The emerging role and clinical significance of circRNAs in papillary thyroid cancer.Front Endocrinol (Lausanne). 2024 Mar 13;15:1351776. doi: 10.3389/fendo.2024.1351776. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38544689 Free PMC article. Review.
Cited by
-
Emerging role of competing endogenous RNA and associated noncoding RNAs in thyroid cancer.Am J Cancer Res. 2022 Mar 15;12(3):961-973. eCollection 2022. Am J Cancer Res. 2022. PMID: 35411240 Free PMC article. Review.
-
Cystic (including atypical) and solid breast lesion classification using the different features of quantitative ultrasound parametric images.Int J Comput Assist Radiol Surg. 2022 Feb;17(2):219-228. doi: 10.1007/s11548-021-02522-x. Epub 2021 Nov 2. Int J Comput Assist Radiol Surg. 2022. PMID: 34727337
-
The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration.Cells. 2023 Jun 14;12(12):1621. doi: 10.3390/cells12121621. Cells. 2023. PMID: 37371091 Free PMC article. Review.
-
CircSEMA6A upregulates PRRG4 by targeting MiR-520h and recruiting ELAVL1 to affect cell invasion and migration in papillary thyroid carcinoma.Arch Endocrinol Metab. 2024 Feb 23;68:e210541. doi: 10.20945/2359-4292-2021-0541. Arch Endocrinol Metab. 2024. PMID: 38394156 Free PMC article.
-
Significance of Circular FAT1 as a Prognostic Factor and Tumor Suppressor for Esophageal Squamous Cell Carcinoma.Ann Surg Oncol. 2021 Dec;28(13):8508-8518. doi: 10.1245/s10434-021-10089-9. Epub 2021 Jun 29. Ann Surg Oncol. 2021. PMID: 34185205 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

