Deep metabolome: Applications of deep learning in metabolomics
- PMID: 33133423
- PMCID: PMC7575644
- DOI: 10.1016/j.csbj.2020.09.033
Deep metabolome: Applications of deep learning in metabolomics
Abstract
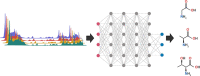
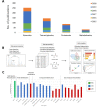
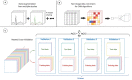
In the past few years, deep learning has been successfully applied to various omics data. However, the applications of deep learning in metabolomics are still relatively low compared to others omics. Currently, data pre-processing using convolutional neural network architecture appears to benefit the most from deep learning. Compound/structure identification and quantification using artificial neural network/deep learning performed relatively better than traditional machine learning techniques, whereas only marginally better results are observed in biological interpretations. Before deep learning can be effectively applied to metabolomics, several challenges should be addressed, including metabolome-specific deep learning architectures, dimensionality problems, and model evaluation regimes.
Keywords: AI, Artificial Intelligence; ANN, Artificial Neural Network; AUC, Area Under the receiver-operating characteristic Curve; Artificial neural network; CCS value, Collision Cross Section value; CFM-EI, Competitive Fragmentation Modeling-Electron Ionization; CNN, Convolutional Neural Network; DL, Deep Learning; DNN, Deep Neural Network; Deep learning; ECFP, Extended Circular Fingerprint; ER, Estrogen Receptor; FID, Free Induction Decay; FP score, Fingerprint correlation score; FTIR, Fourier Transform Infrared; GC–MS, Gas Chromatography-Mass Spectrometry; HDLSS data, High Dimensional Low Sample Size data; IST, Iterative Soft Thresholding; LC-MS, Liquid Chromatography-Mass Spectrometry; LSTM, Long Short-Term Memory; ML, Machine Learning; MLP, Multi-layered Perceptron; MS, Mass Spectrometry; Mass spectrometry; Metabolomics; NEIMS, Neural Electron-Ionization Mass Spectrometry; NMR; NMR, Nuclear Magnetic Resonance; NUS, Non-Uniformly Sampling; PARAFAC2, Parallel Factor Analysis 2; RF, Random Forest; RNN, Recurrent Neural Network; ReLU, Rectified Linear Unit; SMARTS, SMILES arbitrary target specification; SMILE, Sparse Multidimensional Iterative Lineshape-enhanced; SMILES, Simplified Molecular-Input Line-Entry System; SRA, Sequence Read Archive; VAE, Variational Autoencoder; istHMS, Implementation of IST at Harvard Medical School; m/z, mass/charge ratio.
© 2020 The Author(s).
Figures
References
-
- Cambiaghi A., Ferrario M., Masseroli M. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Briefings Bioinf. 2016;18(3):498–510. - PubMed
-
- Smith R., Ventura D., Prince J.T. LC-MS alignment in theory and practice: a comprehensive algorithmic review. Briefings Bioinf. 2013;16(1):104–117. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
