An integrative multi-omics network-based approach identifies key regulators for breast cancer
- PMID: 33133424
- PMCID: PMC7585874
- DOI: 10.1016/j.csbj.2020.10.001
An integrative multi-omics network-based approach identifies key regulators for breast cancer
Abstract
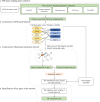
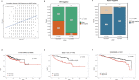
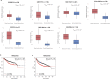
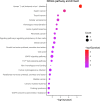
Although genome-wide association studies (GWASs) have successfully identified thousands of risk variants for human complex diseases, understanding the biological function and molecular mechanisms of the associated SNPs involved in complex diseases is challenging. Here we developed a framework named integrative multi-omics network-based approach (IMNA), aiming to identify potential key genes in regulatory networks by integrating molecular interactions across multiple biological scales, including GWAS signals, gene expression-based signatures, chromatin interactions and protein interactions from the network topology. We applied this approach to breast cancer, and prioritized key genes involved in regulatory networks. We also developed an abnormal gene expression score (AGES) signature based on the gene expression deviation of the top 20 rank-ordered genes in breast cancer. The AGES values are associated with genetic variants, tumor properties and patient survival outcomes. Among the top 20 genes, RNASEH2A was identified as a new candidate gene for breast cancer. Thus, our integrative network-based approach provides a genetic-driven framework to unveil tissue-specific interactions from multiple biological scales and reveal potential key regulatory genes for breast cancer. This approach can also be applied in other complex diseases such as ovarian cancer to unravel underlying mechanisms and help for developing therapeutic targets.
Keywords: Breast cancer; GWASs; Integrative network-based approach; Multi-omics; Regulatory network.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J Clinicians. 2018;68(6):394–424. - PubMed
LinkOut - more resources
Full Text Sources
