Genomics of asthma, allergy and chronic rhinosinusitis: novel concepts and relevance in airway mucosa
- PMID: 33133517
- PMCID: PMC7592594
- DOI: 10.1186/s13601-020-00347-6
Genomics of asthma, allergy and chronic rhinosinusitis: novel concepts and relevance in airway mucosa
Abstract
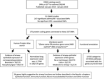
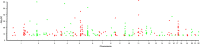
Genome wide association studies (GWASs) have revealed several airway disease-associated risk loci. Their role in the onset of asthma, allergic rhinitis (AR) or chronic rhinosinusitis (CRS), however, is not yet fully understood. The aim of this review is to evaluate the airway relevance of loci and genes identified in GWAS studies. GWASs were searched from databases, and a list of loci associating significantly (p < 10-8) with asthma, AR and CRS was created. This yielded a total of 267 significantly asthma/AR-associated loci from 31 GWASs. No significant CRS -associated loci were found in this search. A total of 170 protein coding genes were connected to these loci. Of these, 76/170 (44%) showed bronchial epithelial protein expression in stained microscopic figures of Human Protein Atlas (HPA), and 61/170 (36%) had a literature report of having airway epithelial function. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation analyses were performed, and 19 functional protein categories were found as significantly (p < 0.05) enriched among these genes. These were related to cytokine production, cell activation and adaptive immune response, and all were strongly connected in network analysis. We also identified 15 protein pathways that were significantly (p < 0.05) enriched in these genes, related to T-helper cell differentiation, virus infection, JAK-STAT signaling pathway, and asthma. A third of GWAS-level risk loci genes of asthma or AR seemed to have airway epithelial functions according to our database and literature searches. In addition, many of the risk loci genes were immunity related. Some risk loci genes also related to metabolism, neuro-musculoskeletal or other functions. Functions overlapped and formed a strong network in our pathway analyses and are worth future studies of biomarker and therapeutics.
Keywords: Airway epithelium; Allergic rhinitis; Asthma; GWAS; Gene ontology; Pathway.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsSTS has acted as paid consultant for Mylan Laboratories Ltd., Biomedical systems Ltd., Roche Products Ltd., and Sanofi S.A. All other authors declare no conflicts of interest.
Figures



Similar articles
-
Airway Epithelial Expression Quantitative Trait Loci Reveal Genes Underlying Asthma and Other Airway Diseases.Am J Respir Cell Mol Biol. 2016 Feb;54(2):177-87. doi: 10.1165/rcmb.2014-0381OC. Am J Respir Cell Mol Biol. 2016. PMID: 26102239 Free PMC article.
-
Multi-omics colocalization with genome-wide association studies reveals a context-specific genetic mechanism at a childhood onset asthma risk locus.Genome Med. 2021 Oct 10;13(1):157. doi: 10.1186/s13073-021-00967-y. Genome Med. 2021. PMID: 34629083 Free PMC article.
-
Sinonasal anatomic variants and asthma are associated with faster development of chronic rhinosinusitis in patients with allergic rhinitis.Int Forum Allergy Rhinol. 2013 Sep;3(9):755-61. doi: 10.1002/alr.21163. Epub 2013 Mar 15. Int Forum Allergy Rhinol. 2013. PMID: 23504927
-
Epigenetics and the Environment in Airway Disease: Asthma and Allergic Rhinitis.Adv Exp Med Biol. 2020;1253:153-181. doi: 10.1007/978-981-15-3449-2_6. Adv Exp Med Biol. 2020. PMID: 32445095 Review.
-
Allergic rhinitis and inflammatory airway disease: interactions within the unified airspace.Am J Rhinol Allergy. 2010 Jul-Aug;24(4):249-54. doi: 10.2500/ajra.2010.24.3499. Am J Rhinol Allergy. 2010. PMID: 20819460 Review.
Cited by
-
Type 2 chronic inflammatory diseases: targets, therapies and unmet needs.Nat Rev Drug Discov. 2023 Sep;22(9):743-767. doi: 10.1038/s41573-023-00750-1. Epub 2023 Aug 1. Nat Rev Drug Discov. 2023. PMID: 37528191 Review.
-
Impact of Interleukin-17 Receptor A Gene Variants on Asthma Susceptibility and Clinical Manifestations in Children and Adolescents.Children (Basel). 2024 May 28;11(6):657. doi: 10.3390/children11060657. Children (Basel). 2024. PMID: 38929236 Free PMC article.
-
Unspecified asthma, childhood-onset, and adult-onset asthma have different causal genes: a Mendelian randomization analysis.Front Immunol. 2024 Nov 19;15:1412032. doi: 10.3389/fimmu.2024.1412032. eCollection 2024. Front Immunol. 2024. PMID: 39628479 Free PMC article.
-
Induction of a type 2 inflammatory chronic rhinosinusitis in C57BL/6 mice.Asia Pac Allergy. 2023 Dec;13(4):164-174. doi: 10.5415/apallergy.0000000000000123. Epub 2023 Nov 2. Asia Pac Allergy. 2023. PMID: 38094094 Free PMC article.
-
GARP Polymorphisms Associated with Susceptibility to House Dust Mite-Sensitized Persistent Allergic Rhinitis in a Chinese Population.J Asthma Allergy. 2022 Sep 28;15:1369-1381. doi: 10.2147/JAA.S366815. eCollection 2022. J Asthma Allergy. 2022. PMID: 36196093 Free PMC article.
References
-
- Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists Rhinology. 2012; 50(1): 1–12 - PubMed
-
- de Loos DD, Lourijsen ES, Wildeman MAM, Freling NJM, Wolvers MDJ, Reitsma S, et al. Prevalence of chronic rhinosinusitis in the general population based on sinus radiology and symptomatology. J Allergy Clin Immunol. 2019; 143(3): 1207–14. - PubMed
-
- Fokkens WJ, Lund VJ, Hopkins C, Hellings PW, Kern R, Reitsma S, et al. European Position Paper on Rhinosinusitis and Nasal Polyps, vol 103. Dieudonné Nyenbue Tshipukane. 2020.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

