A blueprint for the implementation of a validated approach for the detection of SARS-Cov2 in clinical samples in academic facilities
- PMID: 33134554
- PMCID: PMC7590889
- DOI: 10.12688/wellcomeopenres.15937.2
A blueprint for the implementation of a validated approach for the detection of SARS-Cov2 in clinical samples in academic facilities
Abstract
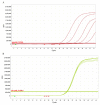
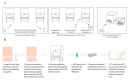
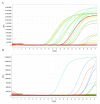
The COVID-19 pandemic is expanding at an unprecedented rate. As a result, diagnostic services are stretched to their limit, and there is a clear need for the provision of additional diagnostic capacity. Academic laboratories, many of which are closed due to governmental lockdowns, may be in a position to support local screening capacity by adapting their current laboratory practices. Here, we describe the process of developing a SARS-Cov2 diagnostic workflow in a conventional academic Containment Level 2 laboratory. Our outline includes simple SARS-Cov2 deactivation upon contact, the method for a quantitative real-time reverse transcriptase PCR detecting SARS-Cov2, a description of process establishment and validation, and some considerations for establishing a similar workflow elsewhere. This was achieved under challenging circumstances through the collaborative efforts of scientists, clinical staff, and diagnostic staff to mitigate to the ongoing crisis. Within 14 days, we created a validated COVID-19 diagnostics service for healthcare workers in our local hospital. The described methods are not exhaustive, but we hope may offer support to other academic groups aiming to set up something comparable in a short time frame.
Keywords: COVID-19; SARS-Cov2; diagnostic PCR; qPCR; sample workflow; validation.
Copyright: © 2020 Sridhar S et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

