Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers
- PMID: 33134617
- PMCID: PMC7584792
- DOI: 10.1016/j.pld.2020.06.003
Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers
Abstract
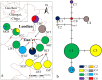
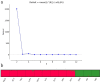
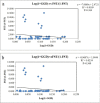
Camellia huana is an endangered species with a narrow distribution in limestone hills of northern Guangxi and southern Guizhou provinces, China. We used one chloroplast DNA (cpDNA) fragment and 12 pairs of microsatellite (simple sequence repeat; SSR) markers to assess the genetic diversity and structure of 12 C. huana populations. A total of 99 alleles were detected for 12 polymorphic loci, and eight haplotypes and nine polymorphic sites were detected within 5200 bp of cpDNA. C. huana populations showed a low level of genetic diversity (n = 8, Hd = 0.759, Pi = 0.00042 for cpDNA, N A = 3.931, H E = 0.466 for SSRs), but high genetic differentiation between populations (F ST = 0.2159 for SSRs, F ST = 0.9318 for cpDNA). This can be attributed to the narrow distribution and limestone habitat of C. huana. STRUCTURE analysis divided natural C. huana populations into two groups, consistent with their geographical distribution. Thus, we suggest that five natural C. huana populations should be split into two units to be managed effectively.
Keywords: AMOVA, analysis of molecular variance; CTAB, cetyl trimethylammonium bromide; Camellia huana; Conservation implications; Genetic diversity; Genetic structure; PCR, polymerase chain reaction; PCoA, principal coordinate analysis; SMM, stepwise mutation model; SSC, small single-copy; SSR, simple sequence repeat; TPM, two-phased model; cpDNA, chloroplast DNA.
© 2020 Kunming Institute of Botany, Chinese Academy of Sciences. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
SSR marker-based genetic diversity and structure analyses of Camellia nitidissima var. phaeopubisperma from different populations.PeerJ. 2025 Jan 21;13:e18845. doi: 10.7717/peerj.18845. eCollection 2025. PeerJ. 2025. PMID: 39866559 Free PMC article.
-
Assessment of the Genetic Relationship and Population Structure in Oil-Tea Camellia Species Using Simple Sequence Repeat (SSR) Markers.Genes (Basel). 2022 Nov 19;13(11):2162. doi: 10.3390/genes13112162. Genes (Basel). 2022. PMID: 36421835 Free PMC article.
-
Comparison of the Chloroplast Genome Sequences of 13 Oil-Tea Camellia Samples and Identification of an Undetermined Oil-Tea Camellia Species From Hainan Province.Front Plant Sci. 2022 Feb 7;12:798581. doi: 10.3389/fpls.2021.798581. eCollection 2021. Front Plant Sci. 2022. PMID: 35197990 Free PMC article.
-
Decoupled mitochondrial and chloroplast DNA population structure reveals Holocene collapse and population isolation in a threatened Mexican-endemic conifer.Mol Ecol. 2006 Sep;15(10):2787-800. doi: 10.1111/j.1365-294X.2006.02974.x. Mol Ecol. 2006. PMID: 16911200
-
Chloroplast DNA phylogeography of Clintoniaudensis Trautv. & Mey. (Liliaceae) in East Asia.Mol Phylogenet Evol. 2010 May;55(2):721-32. doi: 10.1016/j.ympev.2010.02.010. Epub 2010 Feb 19. Mol Phylogenet Evol. 2010. PMID: 20172032 Review.
Cited by
-
Complete Chloroplast Genome Sequence of the Endemic and Endangered Plant Dendropanax oligodontus: Genome Structure, Comparative and Phylogenetic Analysis.Genes (Basel). 2022 Nov 4;13(11):2028. doi: 10.3390/genes13112028. Genes (Basel). 2022. PMID: 36360265 Free PMC article.
-
Population Structure and Genetic Diversity of the Toona ciliata (Meliaceae) Complex Assayed with Chloroplast DNA Markers.Genes (Basel). 2024 Feb 28;15(3):320. doi: 10.3390/genes15030320. Genes (Basel). 2024. PMID: 38540379 Free PMC article.
-
Genetic diversity, population genetic structure and gene flow in the rare and endangered wild plant Cypripedium macranthos revealed by genotyping-by-sequencing.BMC Plant Biol. 2023 May 15;23(1):254. doi: 10.1186/s12870-023-04212-z. BMC Plant Biol. 2023. PMID: 37189068 Free PMC article.
-
Complete Plastome of Physalis angulata var. villosa, Gene Organization, Comparative Genomics and Phylogenetic Relationships among Solanaceae.Genes (Basel). 2022 Dec 5;13(12):2291. doi: 10.3390/genes13122291. Genes (Basel). 2022. PMID: 36553558 Free PMC article.
-
Development of SSR molecular markers and genetic diversity analysis of Clematis acerifolia from Taihang Mountains.PLoS One. 2023 May 19;18(5):e0285754. doi: 10.1371/journal.pone.0285754. eCollection 2023. PLoS One. 2023. PMID: 37205665 Free PMC article.
References
-
- Abbasov M., Akparov Z., Gross T. Genetic relationship of diploid wheat (Triticum spp.) species assessed by SSR markers. Genet. Resour. Crop Evol. 2018;65:1441–1453. doi: 10.1007/s10722-018-0629-2. - DOI
-
- An M.T. Present status of the natural resource of camellias in Guizhou Province. Guizhou Forestry Sci. Tech. 2005;33:26–29.
-
- Bhattacharyya P., Kumaria S. Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplified polymorphic DNA. Plant Systemat. Evol. 2015;301:201–210. doi: 10.1007/s00606-014-1065-1. - DOI
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

