Integrated Micropillar Polydimethylsiloxane Accurate CRISPR Detection System for Viral DNA Sensing
- PMID: 33134706
- PMCID: PMC7594154
- DOI: 10.1021/acsomega.0c03917
Integrated Micropillar Polydimethylsiloxane Accurate CRISPR Detection System for Viral DNA Sensing
Abstract
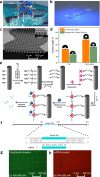
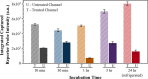
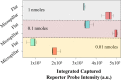
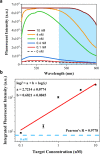
A fully Integrated Micropillar Polydimethylsiloxane Accurate CRISPR deTection (IMPACT) system is developed for viral DNA detection. This powerful system is patterned with high-aspect-ratio micropillars to enhance reporter probe binding. After surface modification and probe immobilization, the CRISPR-Cas12a/crRNA complex is injected into the fully enclosed microchannel. With the presence of a double-stranded DNA target, the CRISPR enzyme is activated and denatures the single-stranded DNA reporters from the micropillars. This collateral cleavage releases fluorescence reporters into the assay, and the intensity is linearly proportional to the target DNA concentration ranging from 0.1 to 10 nM. Importantly, this system does not rely on the traditional dye-quencher-labeled probe, thus reducing the fluorescence background presented in the assay. Furthermore, our one-step detection protocol is performed on-chip at isothermal conditions (37 °C) without using complicated and time-consuming off-chip probe hybridization and denaturation. This miniaturized and fully packed IMPACT chip demonstrates sensitive and accurate DNA detection within 120 min and paves ways to the next-generation point-of-care diagnostics, responding to emerging and deadly pathogen outbreaks.
© 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Coronavirus Update (Live): 18,802,282 Cases and 706,443 Deaths from COVID-19 Virus Pandemic - Worldometer, https://www.worldometers.info/coronavirus/ (accessed Aug 5, 2020).
-
- COVID-19 Map. https://coronavirus.jhu.edu/map.html (accessed Aug 5, 2020).
-
- Cubillos C.; Gómez-Sebastian S.; Moreno N.; Nuñez M. C.; Mulumba-Mfumu L. K.; Quembo C. J.; Heath L.; Etter E. M. C.; Jori F.; Escribano J. M.; Blanco E. African Swine Fever Virus Serodiagnosis: A General Review with a Focus on the Analyses of African Serum Samples. Virus Res. 2013, 173, 159–167. 10.1016/j.virusres.2012.10.021. - DOI - PubMed
-
- Srikrishna D.; Dhillon R. S.; Beier D.. We Need a Cheap Way to Diagnose Coronavirus. Harv. Bus. Rev., 2020.
-
- Borca M. V.; Ramirez-Medina E.; Silva E.; Vuono E.; Rai A.; Pruitt S.; Holinka L. G.; Velazquez-Salinas L.; Zhu J.; Gladue D. P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J. Virol. 2020, 94, e02017 10.1128/JVI.02017-19. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

