Simulating intervertebral disc cell behaviour within 3D multifactorial environments
- PMID: 33135078
- PMCID: PMC8599729
- DOI: 10.1093/bioinformatics/btaa939
Simulating intervertebral disc cell behaviour within 3D multifactorial environments
Abstract
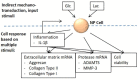
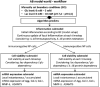
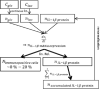
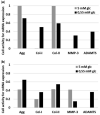
Motivation: Low back pain is responsible for more global disability than any other condition. Its incidence is closely related to intervertebral disc (IVD) failure, which is likely caused by an accumulation of microtrauma within the IVD. Crucial factors in microtrauma development are not entirely known yet, probably because their exploration in vivo or in vitro remains tremendously challenging. In silico modelling is, therefore, definitively appealing, and shall include approaches to integrate influences of multiple cell stimuli at the microscale. Accordingly, this study introduces a hybrid Agent-based (AB) model in IVD research and exploits network modelling solutions in systems biology to mimic the cellular behaviour of Nucleus Pulposus cells exposed to a 3D multifactorial biochemical environment, based on mathematical integrations of existing experimental knowledge. Cellular activity reflected by mRNA expression of Aggrecan, Collagen type I, Collagen type II, MMP-3 and ADAMTS were calculated for inflamed and non-inflamed cells. mRNA expression over long periods of time is additionally determined including cell viability estimations. Model predictions were eventually validated with independent experimental data.
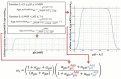
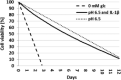
Results: As it combines experimental data to simulate cell behaviour exposed to a multifactorial environment, the present methodology was able to reproduce cell death within 3 days under glucose deprivation and a 50% decrease in cell viability after 7 days in an acidic environment. Cellular mRNA expression under non-inflamed conditions simulated a quantifiable catabolic shift under an adverse cell environment, and model predictions of mRNA expression of inflamed cells provide new explanation possibilities for unexpected results achieved in experimental research.
Availabilityand implementation: The AB model as well as used mathematical functions were built with open source software. Final functions implemented in the AB model and complete AB model parameters are provided as Supplementary Material. Experimental input and validation data were provided through referenced, published papers. The code corresponding to the model can be shared upon request and shall be reused after proper training.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



Similar articles
-
Evidence-Based Network Modelling to Simulate Nucleus Pulposus Multicellular Activity in Different Nutritional and Pro-Inflammatory Environments.Front Bioeng Biotechnol. 2021 Nov 10;9:734258. doi: 10.3389/fbioe.2021.734258. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34858955 Free PMC article. Review.
-
Potential of co-culture of nucleus pulposus mesenchymal stem cells and nucleus pulposus cells in hyperosmotic microenvironment for intervertebral disc regeneration.Cell Biol Int. 2013 Aug;37(8):826-34. doi: 10.1002/cbin.10110. Epub 2013 Apr 18. Cell Biol Int. 2013. PMID: 23554141
-
Immunomodulation of Human Mesenchymal Stem/Stromal Cells in Intervertebral Disc Degeneration: Insights From a Proinflammatory/Degenerative Ex Vivo Model.Spine (Phila Pa 1976). 2018 Jun 15;43(12):E673-E682. doi: 10.1097/BRS.0000000000002494. Spine (Phila Pa 1976). 2018. PMID: 29189572
-
Role of Sirt1 Plays in Nucleus Pulposus Cells and Intervertebral Disc Degeneration.Spine (Phila Pa 1976). 2017 Jul 1;42(13):E757-E766. doi: 10.1097/BRS.0000000000001954. Spine (Phila Pa 1976). 2017. PMID: 27792110
-
Mechanobiology of annulus fibrosus and nucleus pulposus cells in intervertebral discs.Cell Tissue Res. 2020 Mar;379(3):429-444. doi: 10.1007/s00441-019-03136-1. Epub 2019 Dec 17. Cell Tissue Res. 2020. PMID: 31844969 Review.
Cited by
-
Comparison of Clinical and Radiographic Outcomes Between Transforaminal Endoscopic Lumbar Discectomy and Microdiscectomy: A Follow-up Exceeding 5 Years.Neurospine. 2024 Mar;21(1):303-313. doi: 10.14245/ns.2347026.513. Epub 2024 Feb 1. Neurospine. 2024. PMID: 38317550 Free PMC article.
-
Parallel Networks to Predict TIMP and Protease Cell Activity of Nucleus Pulposus Cells Exposed and Not Exposed to Pro-Inflammatory Cytokines.JOR Spine. 2025 Feb 20;8(1):e70051. doi: 10.1002/jsp2.70051. eCollection 2025 Mar. JOR Spine. 2025. PMID: 39981424 Free PMC article.
-
Immuno-Modulatory Effects of Intervertebral Disc Cells.Front Cell Dev Biol. 2022 Jun 29;10:924692. doi: 10.3389/fcell.2022.924692. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35846355 Free PMC article. Review.
-
Investigating the physiological relevance of ex vivo disc organ culture nutrient microenvironments using in silico modeling and experimental validation.JOR Spine. 2021 Mar 2;4(2):e1141. doi: 10.1002/jsp2.1141. eCollection 2021 Jun. JOR Spine. 2021. PMID: 34337330 Free PMC article.
-
In silico modeling the potential clinical effect of growth factor treatment on the metabolism of human nucleus pulposus cells.JOR Spine. 2024 Jul 31;7(3):e1352. doi: 10.1002/jsp2.1352. eCollection 2024 Sep. JOR Spine. 2024. PMID: 39092165 Free PMC article.
References
-
- Baumgartner L. et al. (2019) Simulation of the multifactorial cellular environment within the intervertebral disc to better understand microtrauma emergence. In: Short Communication, International Research Council on the Biomechanics of Injury (IRCOBI) Conference, Florence, Italy, IRC-19-67. pp. 484–485.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

