Meta-analysis based gene expression profiling reveals functional genes in ovarian cancer
- PMID: 33135729
- PMCID: PMC7677829
- DOI: 10.1042/BSR20202911
Meta-analysis based gene expression profiling reveals functional genes in ovarian cancer
Abstract
Background: Ovarian cancer causes high mortality rate worldwide, and despite numerous attempts, the outcome for patients with ovarian cancer are still not well improved. Microarray-based gene expressional analysis provides with valuable information for discriminating functional genes in ovarian cancer development and progression. However, due to the differences in experimental design, the results varied significantly across individual datasets.
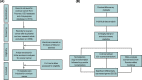
Methods: In the present study, the data of gene expression in ovarian cancer were downloaded from Gene Expression Omnibus (GEO) and 16 studies were included. A meta-analysis based gene expression analysis was performed to identify differentially expressed genes (DEGs). The most differentially expressed genes in our meta-analysis were selected for gene expression and gene function validation.
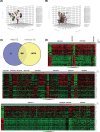
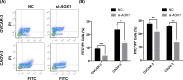
Results: A total of 972 DEGs with P-value < 0.001 were identified in ovarian cancer, including 541 up-regulated genes and 431 down-regulated genes, among which 92 additional DEGs were found as gained DEGs. Top five up- and down-regulated genes were selected for the validation of gene expression profiling. Among these genes, up-regulated CD24 molecule (CD24), SRY (sex determining region Y)-box transcription factor 17 (SOX17), WFDC2, epithelial cell adhesion molecule (EPCAM), innate immunity activator (INAVA), and down-regulated aldehyde oxidase 1 (AOX1) were revealed to be with consistent expressional patterns in clinical patient samples of ovarian cancer. Gene functional analysis demonstrated that up-regulated WFDC2 and INAVA promoted ovarian cancer cell migration, WFDC2 enhanced cell proliferation, while down-regulated AOX1 was functional in inducing cell apoptosis of ovarian cancer.
Conclusion: Our study shed light on the molecular mechanisms underlying the development of ovarian cancer, and facilitated the understanding of novel diagnostic and therapeutic targets in ovarian cancer.
Keywords: Differentially expressed genes; Gene expression omnibus; Microarray analysis; Ovarian cancer.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



Similar articles
-
Cross‑validation of genes potentially associated with neoadjuvant chemotherapy and platinum‑based chemoresistance in epithelial ovarian carcinoma.Oncol Rep. 2020 Sep;44(3):909-926. doi: 10.3892/or.2020.7668. Epub 2020 Jul 2. Oncol Rep. 2020. PMID: 32705213 Free PMC article.
-
Identification of metastasis and prognosis-associated genes for serous ovarian cancer.Biosci Rep. 2020 Jun 26;40(6):BSR20194324. doi: 10.1042/BSR20194324. Biosci Rep. 2020. PMID: 32510146 Free PMC article.
-
Investigation of hypoxia networks in ovarian cancer via bioinformatics analysis.J Ovarian Res. 2018 Feb 26;11(1):16. doi: 10.1186/s13048-018-0388-x. J Ovarian Res. 2018. PMID: 29482638 Free PMC article.
-
DNA microarrays for assessing ovarian cancer gene expression.Mol Cell Endocrinol. 2002 May 31;191(1):121-6. doi: 10.1016/s0303-7207(02)00063-1. Mol Cell Endocrinol. 2002. PMID: 12044926 Review.
-
Overview of the signaling pathways involved in metastasis: An intriguing story-tale of the metastatic journey of ovarian cancer cells.Cell Mol Biol (Noisy-le-grand). 2021 Nov 25;67(3):212-223. doi: 10.14715/cmb/2021.67.3.34. Cell Mol Biol (Noisy-le-grand). 2021. PMID: 34933706 Review.
Cited by
-
The INAVA mRNA in Extracellular Vesicles Activates Normal Ovarian Fibroblasts by Phosphorylation-Ubiquitylation Crosstalk of HMGA2.Adv Sci (Weinh). 2025 Jul;12(25):e2500912. doi: 10.1002/advs.202500912. Epub 2025 Apr 23. Adv Sci (Weinh). 2025. PMID: 40265981 Free PMC article.
-
Integrated Bioinformatics Analysis for Identification of the Hub Genes Linked with Prognosis of Ovarian Cancer Patients.Comput Math Methods Med. 2022 Jan 10;2022:5113447. doi: 10.1155/2022/5113447. eCollection 2022. Comput Math Methods Med. 2022. Retraction in: Comput Math Methods Med. 2023 Dec 6;2023:9826839. doi: 10.1155/2023/9826839. PMID: 35047055 Free PMC article. Retracted.
-
High-grade ovarian cancer associated H/ACA snoRNAs promote cancer cell proliferation and survival.NAR Cancer. 2022 Jan 14;4(1):zcab050. doi: 10.1093/narcan/zcab050. eCollection 2022 Mar. NAR Cancer. 2022. PMID: 35047824 Free PMC article.
-
Investigating the function and targeting of MET protein as an oncogene kinase in pancreatic ductal adenocarcinoma: A microarray data integration.Bioimpacts. 2024 Oct 1;15:30187. doi: 10.34172/bi.30187. eCollection 2025. Bioimpacts. 2024. PMID: 40161938 Free PMC article. Review.
-
Comprehensively analyzing the genetic alterations, and identifying key genes in ovarian cancer.Oncol Res. 2023 Apr 10;31(2):141-156. doi: 10.32604/or.2023.028548. eCollection 2023. Oncol Res. 2023. PMID: 37304238 Free PMC article.
References
-
- Urban N. and Drescher C. (2006) Current and future developments in screening for ovarian cancer. Womens Health (Lond.) 2, 733–742 - PubMed
-
- Bowen N.J., Walker L.D., Matyunina L.V., Logani S., Totten K.A., Benigno B.B. et al. . (2009) Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med. Genomics 2, 71 10.1186/1755-8794-2-71 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

