Functional and druggability analysis of the SARS-CoV-2 proteome
- PMID: 33137330
- PMCID: PMC7604074
- DOI: 10.1016/j.ejphar.2020.173705
Functional and druggability analysis of the SARS-CoV-2 proteome
Abstract
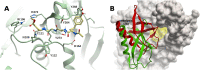
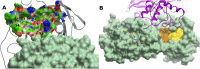
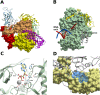
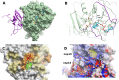
The infectious coronavirus disease (COVID-19) pandemic, caused by the coronavirus SARS-CoV-2, appeared in December 2019 in Wuhan, China, and has spread worldwide. As of today, more than 46 million people have been infected and over 1.2 million fatalities. With the purpose of contributing to the development of effective therapeutics, we performed an in silico determination of binding hot-spots and an assessment of their druggability within the complete SARS-CoV-2 proteome. All structural, non-structural, and accessory proteins have been studied, and whenever experimental structural data of SARS-CoV-2 proteins were not available, homology models were built based on solved SARS-CoV structures. Several potential allosteric or protein-protein interaction druggable sites on different viral targets were identified, knowledge that could be used to expand current drug discovery endeavors beyond the currently explored cysteine proteases and the polymerase complex. It is our hope that this study will support the efforts of the scientific community both in understanding the molecular determinants of this disease and in widening the repertoire of viral targets in the quest for repurposed or novel drugs against COVID-19.
Keywords: Binding hot-spots; COVID-19; Coronavirus; Drug discovery; Druggability; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Figures






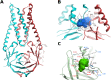
Similar articles
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
In silico Drug Repurposing for COVID-19: Targeting SARS-CoV-2 Proteins through Docking and Consensus Ranking.Mol Inform. 2021 Jan;40(1):e2000115. doi: 10.1002/minf.202000115. Epub 2020 Aug 18. Mol Inform. 2021. PMID: 32722864
-
Molecular mechanisms of the novel coronavirus SARS-CoV-2 and potential anti-COVID19 pharmacological targets since the outbreak of the pandemic.Food Chem Toxicol. 2020 Dec;146:111805. doi: 10.1016/j.fct.2020.111805. Epub 2020 Oct 8. Food Chem Toxicol. 2020. PMID: 33038452 Free PMC article. Review.
-
Mapping major SARS-CoV-2 drug targets and assessment of druggability using computational fragment screening: Identification of an allosteric small-molecule binding site on the Nsp13 helicase.PLoS One. 2021 Feb 17;16(2):e0246181. doi: 10.1371/journal.pone.0246181. eCollection 2021. PLoS One. 2021. PMID: 33596235 Free PMC article.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
Cited by
-
Identification of 1H-purine-2,6-dione derivative as a potential SARS-CoV-2 main protease inhibitor: molecular docking, dynamic simulations, and energy calculations.PeerJ. 2022 Oct 7;10:e14120. doi: 10.7717/peerj.14120. eCollection 2022. PeerJ. 2022. PMID: 36225900 Free PMC article.
-
An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2.Comput Struct Biotechnol J. 2021;19:4868-4883. doi: 10.1016/j.csbj.2021.08.036. Epub 2021 Aug 24. Comput Struct Biotechnol J. 2021. PMID: 34457214 Free PMC article. Review.
-
Lipid polymer hybrid nanocarriers as a combinatory platform for different anti-SARS-CoV-2 drugs supported by computational studies.RSC Adv. 2021 Aug 27;11(46):28876-28891. doi: 10.1039/d1ra04576h. eCollection 2021 Aug 23. RSC Adv. 2021. PMID: 35478590 Free PMC article.
-
The importance of accessory protein variants in the pathogenicity of SARS-CoV-2.Arch Biochem Biophys. 2022 Mar 15;717:109124. doi: 10.1016/j.abb.2022.109124. Epub 2022 Jan 24. Arch Biochem Biophys. 2022. PMID: 35085577 Free PMC article.
-
Resources and computational strategies to advance small molecule SARS-CoV-2 discovery: Lessons from the pandemic and preparing for future health crises.Comput Struct Biotechnol J. 2021;19:2537-2548. doi: 10.1016/j.csbj.2021.04.059. Epub 2021 Apr 26. Comput Struct Biotechnol J. 2021. PMID: 33936562 Free PMC article. Review.
References
-
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 218. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

