Human coronavirus spike protein-host receptor recognition
- PMID: 33137344
- PMCID: PMC7604128
- DOI: 10.1016/j.pbiomolbio.2020.10.006
Human coronavirus spike protein-host receptor recognition
Abstract
A variety of coronaviruses (CoVs) have infected humans and caused mild to severe respiratory diseases that could result in mortality. The human CoVs (HCoVs) belong to the genera of α- and β-CoVs that originate in rodents and bats and are transmitted to humans via zoonotic contacts. The binding of viral spike proteins to the host cell receptors is essential for mediating fusion of viral and host cell membranes to cause infection. The SARS-CoV-2 originated in bats (RaTG13 SARS-CoV) and is transmitted to humans via pangolins. The presence of 'PRRA' sequence motif in SARS-CoV-2 spike proteins from human, dog, cat, mink, tiger and lion suggests a common viral entry mechanism into host cells. In this review, we discuss structural features of HCoV spike proteins and recognition of host protein and carbohydrate receptors.
Keywords: Amino peptidase N; Angiotensin-converting enzyme 2; Dipeptidyl peptidase 4; HCoV-229E; HCoV-HKU1; HCoV-NL63; HCoV-OC43; Human coronavirus; MERS-CoV; Receptor binding domain; SARS-CoV; SARS-CoV-2; Sialic acid; Spike protein.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The author declares that there is no potential conflict of interest.
Figures



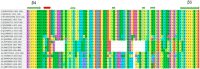
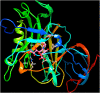

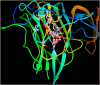

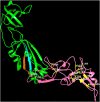


Similar articles
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Characterization of spike S1/S2 processing and entry pathways of lentiviral pseudoviruses bearing seasonal human coronaviruses NL63, 229E, and HKU1 spikes.Microbiol Spectr. 2025 Mar 4;13(3):e0280824. doi: 10.1128/spectrum.02808-24. Epub 2025 Jan 28. Microbiol Spectr. 2025. PMID: 39873512 Free PMC article.
-
Untangling the Evolution of the Receptor-Binding Motif of SARS-CoV-2.J Mol Evol. 2024 Jun;92(3):329-337. doi: 10.1007/s00239-024-10175-y. Epub 2024 May 22. J Mol Evol. 2024. PMID: 38777906 Free PMC article.
-
Structural proteins of human coronaviruses: what makes them different?Front Cell Infect Microbiol. 2024 Dec 6;14:1458383. doi: 10.3389/fcimb.2024.1458383. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39711780 Free PMC article. Review.
-
Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History.J Virol. 2017 Feb 14;91(5):e01953-16. doi: 10.1128/JVI.01953-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28077633 Free PMC article.
Cited by
-
Machine Learning and Artificial Intelligence for the Prediction of Host-Pathogen Interactions: A Viral Case.Infect Drug Resist. 2021 Aug 20;14:3319-3326. doi: 10.2147/IDR.S292743. eCollection 2021. Infect Drug Resist. 2021. PMID: 34456575 Free PMC article. Review.
-
Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases.Cell Commun Signal. 2023 May 15;21(1):110. doi: 10.1186/s12964-023-01104-5. Cell Commun Signal. 2023. PMID: 37189112 Free PMC article. Review.
-
More tools for our toolkit: The application of HEL-299 cells and dsRNA-nanoparticles to study human coronaviruses in vitro.Virus Res. 2022 Nov;321:198925. doi: 10.1016/j.virusres.2022.198925. Epub 2022 Sep 15. Virus Res. 2022. PMID: 36115551 Free PMC article.
-
Taking stock of the mutations in human SARS-CoV-2 spike proteins: From early days to nearly the end of COVID-19 pandemic.Curr Res Struct Biol. 2023 Oct 5;6:100107. doi: 10.1016/j.crstbi.2023.100107. eCollection 2023. Curr Res Struct Biol. 2023. PMID: 37841365 Free PMC article.
-
Prediction of SARS-CoV-2 hosts among Brazilian mammals and new coronavirus transmission chain using evolutionary bioinformatics.Anim Dis. 2021;1(1):20. doi: 10.1186/s44149-021-00020-w. Epub 2021 Sep 26. Anim Dis. 2021. PMID: 34778882 Free PMC article.
References
-
- Almeida J.D., Berry D.M., Cunningham C.H., Hamre D., Hofstad M.S., Mallucci L., McIntosh K., Tyrrell D.A.J. Coronaviruses. Nature. 1968;220(5168):650.
-
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370(26):2499–2505. - PubMed
-
- Azhar E.I., Hashem A.M., El-Kafrawy S.A., Sohrab S.S., Aburizaiza A.S., Farraj S.A., Hassan A.M., Al-Saeed M.S., Jamjoom G.A., Madani T.A. Detection of the Middle East respiratory syndrome coronavirus genome in an air sample originating from a camel barn owned by an infected patient. mBio. 2014;5(4):e01450–14. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

