crAssphage genomes identified in fecal samples of an adult and infants with evidence of positive genomic selective pressure within tail protein genes
- PMID: 33137401
- PMCID: PMC7778472
- DOI: 10.1016/j.virusres.2020.198219
crAssphage genomes identified in fecal samples of an adult and infants with evidence of positive genomic selective pressure within tail protein genes
Abstract
crAssphages are a broad group of diverse bacteriophages in the order Caudovirales that have been found to be highly abundant in the human gastrointestinal tract. Despite their high prevalence, we have an incomplete understanding of how crAssphages shape and respond to ecological and evolutionary dynamics in the gut. Here, we report genomes of crAssphages from feces of one South African woman and three infants. Across the complete genome sequences of the South African crAssphages described here, we identify particularly elevated positive selection in RNA polymerase and phage tail protein encoding genes, contrasted against purifying selection, genome-wide. We further validate these findings against a crAssphage genome from previous studies. Together, our results suggest hotspots of selection within crAssphage RNA polymerase and phage tail protein encoding genes are potentially mediated by interactions between crAssphages and their bacterial partners.
Keywords: Bacteriophage; Selection; South Africa; Tail protein; crAssphage.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures
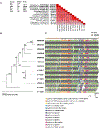
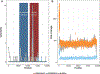
Similar articles
-
Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut.Cell Host Microbe. 2018 Nov 14;24(5):653-664.e6. doi: 10.1016/j.chom.2018.10.002. Epub 2018 Oct 25. Cell Host Microbe. 2018. PMID: 30449316
-
CrAssphage distribution analysis in an Amazonian river based on metagenomic sequencing data and georeferencing.Appl Environ Microbiol. 2025 May 21;91(5):e0147024. doi: 10.1128/aem.01470-24. Epub 2025 Apr 25. Appl Environ Microbiol. 2025. PMID: 40277368 Free PMC article.
-
CrAssphage and its bacterial host in cat feces.Sci Rep. 2021 Jan 12;11(1):815. doi: 10.1038/s41598-020-80076-9. Sci Rep. 2021. PMID: 33436756 Free PMC article.
-
CrAss-Like Phages: From Discovery in Human Fecal Metagenome to Application as a Microbial Source Tracking Marker.Food Environ Virol. 2024 Jun;16(2):121-135. doi: 10.1007/s12560-024-09584-5. Epub 2024 Feb 27. Food Environ Virol. 2024. PMID: 38413544 Review.
-
Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae.Res Microbiol. 2008 Jun;159(5):340-8. doi: 10.1016/j.resmic.2008.04.005. Epub 2008 Apr 30. Res Microbiol. 2008. PMID: 18550341 Free PMC article. Review.
Cited by
-
Characterization of crAss-like phage isolates highlights Crassvirales genetic heterogeneity and worldwide distribution.Nat Commun. 2023 Jul 18;14(1):4295. doi: 10.1038/s41467-023-40098-z. Nat Commun. 2023. PMID: 37463935 Free PMC article.
-
Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee.Arch Virol. 2023 Jan 23;168(2):74. doi: 10.1007/s00705-022-05694-2. Arch Virol. 2023. PMID: 36683075 Free PMC article.
-
Virome Characterization in Commercial Bovine Serum Batches-A Potentially Needed Testing Strategy for Biological Products.Viruses. 2021 Dec 3;13(12):2425. doi: 10.3390/v13122425. Viruses. 2021. PMID: 34960693 Free PMC article.
-
Infant gut DNA bacteriophage strain persistence during the first 3 years of life.Cell Host Microbe. 2024 Jan 10;32(1):35-47.e6. doi: 10.1016/j.chom.2023.11.015. Epub 2023 Dec 13. Cell Host Microbe. 2024. PMID: 38096814 Free PMC article.
-
Bacteriophages of the Order Crassvirales: What Do We Currently Know about This Keystone Component of the Human Gut Virome?Biomolecules. 2023 Mar 24;13(4):584. doi: 10.3390/biom13040584. Biomolecules. 2023. PMID: 37189332 Free PMC article. Review.
References
-
- Angly F, Youle M, Nosrat B, Srinagesh S, Rodriguez-Brito B, McNairnie P, Deyanat-Yazdi G, Breitbart M, Rohwer F, 2009. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol 11(11), 2863–2873. - PubMed
-
- Brockhurst MA, Morgan AD, Fenton A, Buckling A, 2007. Experimental coevolution with bacteria and phage. The Pseudomonas fluorescens--Phi2 model system. Infect Genet Evol 7(4), 547–552. - PubMed
-
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM, 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2), 80–92. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

