Genome Size Evolution Mediated by Gypsy Retrotransposons in Brassicaceae
- PMID: 33137519
- PMCID: PMC7801240
- DOI: 10.1016/j.gpb.2018.07.009
Genome Size Evolution Mediated by Gypsy Retrotransposons in Brassicaceae
Abstract
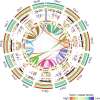
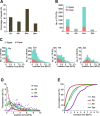
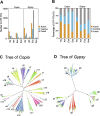
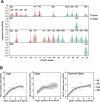
The dynamic activity of transposable elements (TEs) contributes to the vast diversity of genome size and architecture among plants. Here, we examined the genomic distribution and transposition activity of long terminal repeat retrotransposons (LTR-RTs) in Arabidopsis thaliana (Ath) and three of its relatives, Arabidopsis lyrata (Aly), Eutrema salsugineum (Esa), and Schrenkiella parvula (Spa), in Brassicaceae. Our analyses revealed the distinct evolutionary dynamics of Gypsyretrotransposons, which reflects the different patterns of genome size changes of the four species over the past million years. The rate of Gypsy transposition in Aly is approximately five times more rapid than that of Ath and Esa, suggesting an expanding Aly genome. Gypsy insertions in Esa are strictly confined to pericentromeric heterochromatin and associated with dramatic centromere expansion. In contrast, Gypsy insertions in Spa have been largely suppressed over the last million years, likely as a result of a combination of an inherent molecular mechanism of preferential DNA removal and purifying selection at Gypsy elements. Additionally, species-specific clades of Gypsy elements shaped the distinct genome architectures of Aly and Esa.
Keywords: Brassicaceae; Comparative genomics; Copia retrotransposon; Genome size evolution; Gypsy retrotransposon.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
New Insights into Long Terminal Repeat Retrotransposons in Mulberry Species.Genes (Basel). 2019 Apr 9;10(4):285. doi: 10.3390/genes10040285. Genes (Basel). 2019. PMID: 30970574 Free PMC article.
-
Genomic abundance and transcriptional activity of diverse gypsy and copia long terminal repeat retrotransposons in three wild sunflower species.BMC Plant Biol. 2018 Jan 5;18(1):6. doi: 10.1186/s12870-017-1223-z. BMC Plant Biol. 2018. PMID: 29304730 Free PMC article.
-
Mollusc genomes reveal variability in patterns of LTR-retrotransposons dynamics.BMC Genomics. 2018 Nov 15;19(1):821. doi: 10.1186/s12864-018-5200-1. BMC Genomics. 2018. PMID: 30442098 Free PMC article.
-
LTR-retrotransposons in plants: Engines of evolution.Gene. 2017 Aug 30;626:14-25. doi: 10.1016/j.gene.2017.04.051. Epub 2017 May 2. Gene. 2017. PMID: 28476688 Review.
-
Retrotransposons represent the most labile fraction for genomic rearrangements in polyploid plant species.Cytogenet Genome Res. 2013;140(2-4):286-94. doi: 10.1159/000353308. Epub 2013 Jul 9. Cytogenet Genome Res. 2013. PMID: 23899810 Review.
Cited by
-
Novel Insights into the Nature of Intraspecific Genome Size Diversity in Cannabis sativa L.Plants (Basel). 2022 Oct 16;11(20):2736. doi: 10.3390/plants11202736. Plants (Basel). 2022. PMID: 36297761 Free PMC article.
-
Transposable elements in Rosaceae: insights into genome evolution, expression dynamics, and syntenic gene regulation.Hortic Res. 2024 Apr 26;11(6):uhae118. doi: 10.1093/hr/uhae118. eCollection 2024 Jun. Hortic Res. 2024. PMID: 38919560 Free PMC article.
-
Evolutionary history of magnoliid genomes and benzylisoquinoline alkaloid biosynthesis.Nat Commun. 2025 Apr 29;16(1):4039. doi: 10.1038/s41467-025-59343-8. Nat Commun. 2025. PMID: 40301376 Free PMC article.
-
Assembled and annotated 26.5 Gbp coast redwood genome: a resource for estimating evolutionary adaptive potential and investigating hexaploid origin.G3 (Bethesda). 2022 Jan 4;12(1):jkab380. doi: 10.1093/g3journal/jkab380. G3 (Bethesda). 2022. PMID: 35100403 Free PMC article.
-
Transposon dynamics in the emerging oilseed crop Thlaspi arvense.PLoS Genet. 2024 Jan 31;20(1):e1011141. doi: 10.1371/journal.pgen.1011141. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38295109 Free PMC article.
References
-
- Kumar A., Bennetzen J.L. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

