The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing
- PMID: 33137924
- PMCID: PMC7693646
- DOI: 10.3390/foods9111568
The Microbial Diversity of Non-Korean Kimchi as Revealed by Viable Counting and Metataxonomic Sequencing
Abstract
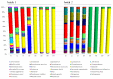
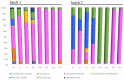
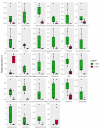
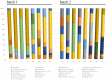
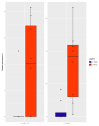
Kimchi is recognized worldwide as the flagship food of Korea. To date, most of the currently available microbiological studies on kimchi deal with Korean manufactures. Moreover, there is a lack of knowledge on the occurrence of eumycetes in kimchi. Given these premises, the present study was aimed at investigating the bacterial and fungal dynamics occurring during the natural fermentation of an artisan non-Korean kimchi manufacture. Lactic acid bacteria were dominant, while Enterobacteriaceae, Pseudomonadaceae, and yeasts progressively decreased during fermentation. Erwinia spp., Pseudomonasveronii, Pseudomonasviridiflava, Rahnellaaquatilis, and Sphingomonas spp. were detected during the first 15 days of fermentation, whereas the last fermentation phase was dominated by Leuconostoc kimchi, together with Weissellasoli. For the mycobiota at the beginning of the fermentation process, Rhizoplaca and Pichia orientalis were the dominant Operational Taxonomic Units (OTUs) in batch 1, whereas in batch 2 Protomyces inundatus prevailed. In the last stage of fermentation, Saccharomyces cerevisiae, Candida sake,Penicillium, and Malassezia were the most abundant taxa in both analyzed batches. The knowledge gained in the present study represents a step forward in the description of the microbial dynamics of kimchi produced outside the region of origin using local ingredients. It will also serve as a starting point for further isolation of kimchi-adapted microorganisms to be assayed as potential starters for the manufacturing of novel vegetable preserves with high quality and functional traits.
Keywords: Candida sake; Leuconostoc kimchii; Weissella soli; fermented vegetables; microbial diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Effects of Leuconostoc mesenteroides starter cultures on microbial communities and metabolites during kimchi fermentation.Int J Food Microbiol. 2012 Feb 15;153(3):378-87. doi: 10.1016/j.ijfoodmicro.2011.11.030. Epub 2011 Dec 4. Int J Food Microbiol. 2012. PMID: 22189023
-
Unraveling microbial fermentation features in kimchi: from classical to meta-omics approaches.Appl Microbiol Biotechnol. 2020 Sep;104(18):7731-7744. doi: 10.1007/s00253-020-10804-8. Epub 2020 Aug 4. Appl Microbiol Biotechnol. 2020. PMID: 32749526 Review.
-
Role of combinated lactic acid bacteria in bacterial, viral, and metabolite dynamics during fermentation of vegetable food, kimchi.Food Res Int. 2022 Jul;157:111261. doi: 10.1016/j.foodres.2022.111261. Epub 2022 Apr 18. Food Res Int. 2022. PMID: 35761573
-
Characterization of starter kimchi fermented with Leuconostoc kimchii GJ2 and its cholesterol-lowering effects in rats fed a high-fat and high-cholesterol diet.J Sci Food Agric. 2015 Oct;95(13):2750-6. doi: 10.1002/jsfa.7018. Epub 2014 Dec 18. J Sci Food Agric. 2015. PMID: 25425317
-
Kimchi microflora: history, current status, and perspectives for industrial kimchi production.Appl Microbiol Biotechnol. 2014 Mar;98(6):2385-93. doi: 10.1007/s00253-014-5513-1. Epub 2014 Jan 14. Appl Microbiol Biotechnol. 2014. PMID: 24419800 Review.
Cited by
-
Growth of ɣ-Proteobacteria in Low Salt Cucumber Fermentation Is Prevented by Lactobacilli and the Cover Brine Ingredients.Microbiol Spectr. 2022 Jun 29;10(3):e0103121. doi: 10.1128/spectrum.01031-21. Epub 2022 May 11. Microbiol Spectr. 2022. PMID: 35543556 Free PMC article.
-
Real-Time Monitoring the Effects of Storage Conditions on Volatile Compounds and Quality Indexes of Halal-Certified Kimchi during Distribution Using Electronic Nose.Foods. 2022 Aug 3;11(15):2323. doi: 10.3390/foods11152323. Foods. 2022. PMID: 35954088 Free PMC article.
-
Vegetable By-Products from Industrial Processing: From Waste to Functional Ingredient Through Fermentation.Foods. 2025 Jul 31;14(15):2704. doi: 10.3390/foods14152704. Foods. 2025. PMID: 40807640 Free PMC article. Review.
-
Knot formation and spread along the shoot stem in 13 olive cultivars inoculated with an indigenous pathobiome of 7 species of Pseudomonas including Pseudomonas savastanoi.PLoS One. 2023 Aug 11;18(8):e0289875. doi: 10.1371/journal.pone.0289875. eCollection 2023. PLoS One. 2023. PMID: 37566625 Free PMC article.
-
Impact of Different Drying Methods on the Microbiota, Volatilome, Color, and Sensory Traits of Sea Fennel (Crithmum maritimum L.) Leaves.Molecules. 2023 Oct 21;28(20):7207. doi: 10.3390/molecules28207207. Molecules. 2023. PMID: 37894688 Free PMC article.
References
-
- Misihairabgwi J., Cheikhyoussef A. Traditional fermented foods and beverages of Namibia. J. Ethn. Foods. 2017;4:145–153. doi: 10.1016/j.jef.2017.08.001. - DOI
-
- Ramos C.L., Schwan R.F. Technological and nutritional aspects of indigenous Latin America fermented foods. Curr. Opin. Food Sci. 2017;13:97–102. doi: 10.1016/j.cofs.2017.07.001. - DOI
-
- Jaehae L., Kyeongsoon H., Chaelin P., Ilgwon K., Jeongwon K., Dukno Y., Montanari M., Naomichi I., Cwiertka K.J., Younghee S., et al. Humanistic Understanding of Kimchi and Kimjang Culture. 1st ed. World Institute of Kimchi; Gwangju, Korea: 2015.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

