Atg43 tethers isolation membranes to mitochondria to promote starvation-induced mitophagy in fission yeast
- PMID: 33138913
- PMCID: PMC7609059
- DOI: 10.7554/eLife.61245
Atg43 tethers isolation membranes to mitochondria to promote starvation-induced mitophagy in fission yeast
Abstract
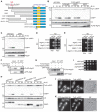
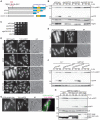
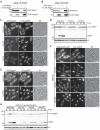
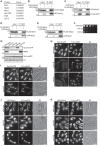
Degradation of mitochondria through mitophagy contributes to the maintenance of mitochondrial function. In this study, we identified that Atg43, a mitochondrial outer membrane protein, serves as a mitophagy receptor in the model organism Schizosaccharomyces pombe to promote the selective degradation of mitochondria. Atg43 contains an Atg8-family-interacting motif essential for mitophagy. Forced recruitment of Atg8 to mitochondria restores mitophagy in Atg43-deficient cells, suggesting that Atg43 tethers expanding isolation membranes to mitochondria. We found that the mitochondrial import factors, including the Mim1-Mim2 complex and Tom70, are crucial for mitophagy. Artificial mitochondrial loading of Atg43 bypasses the requirement of the import factors, suggesting that they contribute to mitophagy through Atg43. Atg43 not only maintains growth ability during starvation but also facilitates vegetative growth through its mitophagy-independent function. Thus, Atg43 is a useful model to study the mechanism and physiological roles, as well as the origin and evolution, of mitophagy in eukaryotes.
Keywords: Atg43; MIM complex; S. pombe; autophagy; cell biology; mitochondria; mitophagy; receptor.
© 2020, Fukuda et al.
Conflict of interest statement
TF, YE, TS, KF, SY, KI, DK, YY, TK No competing interests declared
Figures








Comment in
-
Atg43, a novel autophagy-related protein, serves as a mitophagy receptor to bridge mitochondria with phagophores in fission yeast.Autophagy. 2021 Mar;17(3):826-827. doi: 10.1080/15548627.2021.1874662. Epub 2021 Jan 21. Autophagy. 2021. PMID: 33475472 Free PMC article.
References
-
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. - DOI - PubMed
-
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N. Biogenesis of the mitochondrial TOM complex: mim1 promotes insertion and assembly of signal-anchored receptors. The Journal of Biological Chemistry. 2008;283:120–127. doi: 10.1074/jbc.M706997200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 17K07330/Japan Society for the Promotion of Science/International
- 20K06552/Japan Society for the Promotion of Science/International
- 18H04858/Japan Society for the Promotion of Science/International
- 19H05712/Japan Society for the Promotion of Science/International
- 19K22419/Japan Society for the Promotion of Science/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

