Targeting the Achilles' Heel of Bacteria: Different Mechanisms To Break Down the Peptidoglycan Cell Wall during Bacterial Warfare
- PMID: 33139480
- PMCID: PMC8088523
- DOI: 10.1128/JB.00478-20
Targeting the Achilles' Heel of Bacteria: Different Mechanisms To Break Down the Peptidoglycan Cell Wall during Bacterial Warfare
Abstract
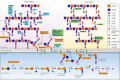
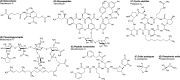
Bacteria commonly live in dense polymicrobial communities and compete for scarce resources. Consequently, they employ a diverse array of mechanisms to harm, inhibit, and kill their competitors. The cell wall is essential for bacterial survival by providing mechanical strength to resist osmotic stress. Because peptidoglycan is the major component of the cell wall and its synthesis is a complex multistep pathway that requires the coordinate action of several enzymes, it provides a target for rival bacteria, which have developed a large arsenal of antibacterial molecules to attack the peptidoglycan of competitors. These molecules include antibiotics, bacteriocins, and contact-dependent effectors that are either secreted into the medium or directly translocated into a target cell. In this minireview, we summarize the diversity of these molecules and highlight distinct mechanisms to disrupt the peptidoglycan, giving special attention to molecules that are known or have the potential to be used during interbacterial competitions.
Keywords: antibiotic; antimicrobial peptide; bacterial warfare; bacteriocin; effector; interbacterial competition; microbial ecology; peptidoglycan.
Copyright © 2021 American Society for Microbiology.
Figures

References
-
- Walsh C. 2003. Antibiotics: actions, origins, resistance. American Society for Microbiology, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

