APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer's disease patient iPSC-derived cerebral organoids
- PMID: 33139712
- PMCID: PMC7608683
- DOI: 10.1038/s41467-020-19264-0
APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer's disease patient iPSC-derived cerebral organoids
Erratum in
-
Author Correction: APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer's disease patient iPSC-derived cerebral organoids.Nat Commun. 2021 May 5;12(1):2707. doi: 10.1038/s41467-021-23081-4. Nat Commun. 2021. PMID: 33953196 Free PMC article. No abstract available.
Abstract
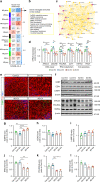
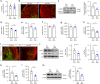
APOE4 is the strongest genetic risk factor associated with late-onset Alzheimer's disease (AD). To address the underlying mechanism, we develop cerebral organoid models using induced pluripotent stem cells (iPSCs) with APOE ε3/ε3 or ε4/ε4 genotype from individuals with either normal cognition or AD dementia. Cerebral organoids from AD patients carrying APOE ε4/ε4 show greater apoptosis and decreased synaptic integrity. While AD patient-derived cerebral organoids have increased levels of Aβ and phosphorylated tau compared to healthy subject-derived cerebral organoids, APOE4 exacerbates tau pathology in both healthy subject-derived and AD patient-derived organoids. Transcriptomics analysis by RNA-sequencing reveals that cerebral organoids from AD patients are associated with an enhancement of stress granules and disrupted RNA metabolism. Importantly, isogenic conversion of APOE4 to APOE3 attenuates the APOE4-related phenotypes in cerebral organoids from AD patients. Together, our study using human iPSC-organoids recapitulates APOE4-related phenotypes and suggests APOE4-related degenerative pathways contributing to AD pathogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures
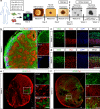
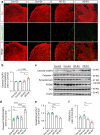
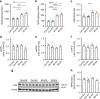
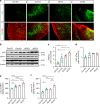

Comment in
-
Brain organoids shed light on APOE genotype and Alzheimer disease pathology.Nat Rev Neurol. 2021 Jan;17(1):1. doi: 10.1038/s41582-020-00437-w. Nat Rev Neurol. 2021. PMID: 33219337 No abstract available.
Similar articles
-
APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer's Disease Phenotypes in Human iPSC-Derived Brain Cell Types.Neuron. 2018 Jun 27;98(6):1141-1154.e7. doi: 10.1016/j.neuron.2018.05.008. Epub 2018 May 31. Neuron. 2018. PMID: 29861287 Free PMC article.
-
APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes.Hum Mol Genet. 2017 Jul 15;26(14):2690-2700. doi: 10.1093/hmg/ddx155. Hum Mol Genet. 2017. PMID: 28444230 Free PMC article.
-
Culture Variabilities of Human iPSC-Derived Cerebral Organoids Are a Major Issue for the Modelling of Phenotypes Observed in Alzheimer's Disease.Stem Cell Rev Rep. 2022 Feb;18(2):718-731. doi: 10.1007/s12015-021-10147-5. Epub 2021 Mar 16. Stem Cell Rev Rep. 2022. PMID: 33725267
-
ApoE4 reduction: An emerging and promising therapeutic strategy for Alzheimer's disease.Neurobiol Aging. 2022 Jul;115:20-28. doi: 10.1016/j.neurobiolaging.2022.03.011. Epub 2022 Mar 22. Neurobiol Aging. 2022. PMID: 35453035 Free PMC article. Review.
-
Apolipoprotein E isoforms in Alzheimer's disease pathology and etiology.Microsc Res Tech. 2000 Aug 15;50(4):278-81. doi: 10.1002/1097-0029(20000815)50:4<278::AID-JEMT5>3.0.CO;2-T. Microsc Res Tech. 2000. PMID: 10936880 Review.
Cited by
-
Human pluripotent stem cell-derived brain organoids as in vitro models for studying neural disorders and cancer.Cell Biosci. 2021 May 28;11(1):99. doi: 10.1186/s13578-021-00617-1. Cell Biosci. 2021. PMID: 34049587 Free PMC article. Review.
-
Lipoprotein Metabolism, Protein Aggregation, and Alzheimer's Disease: A Literature Review.Int J Mol Sci. 2023 Feb 2;24(3):2944. doi: 10.3390/ijms24032944. Int J Mol Sci. 2023. PMID: 36769268 Free PMC article. Review.
-
Protocol for controlled cortical impact in human cerebral organoids to model traumatic brain injury.STAR Protoc. 2021 Dec 4;2(4):100987. doi: 10.1016/j.xpro.2021.100987. eCollection 2021 Dec 17. STAR Protoc. 2021. PMID: 34927096 Free PMC article.
-
APOE4-promoted gliosis and degeneration in tauopathy are ameliorated by pharmacological inhibition of HMGB1 release.Cell Rep. 2023 Oct 31;42(10):113252. doi: 10.1016/j.celrep.2023.113252. Epub 2023 Oct 19. Cell Rep. 2023. PMID: 37863057 Free PMC article.
-
Looking at Alzheimer's Disease Pathogenesis from the Nuclear Side.Biomolecules. 2021 Aug 24;11(9):1261. doi: 10.3390/biom11091261. Biomolecules. 2021. PMID: 34572474 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

