Culicidae evolutionary history focusing on the Culicinae subfamily based on mitochondrial phylogenomics
- PMID: 33139764
- PMCID: PMC7606482
- DOI: 10.1038/s41598-020-74883-3
Culicidae evolutionary history focusing on the Culicinae subfamily based on mitochondrial phylogenomics
Abstract
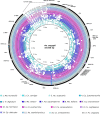
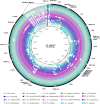
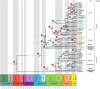
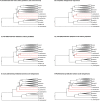
Mosquitoes are insects of medical importance due their role as vectors of different pathogens to humans. There is a lack of information about the evolutionary history and phylogenetic positioning of the majority of mosquito species. Here we characterized the mitogenomes of mosquito species through low-coverage whole genome sequencing and data mining. A total of 37 draft mitogenomes of different species were assembled from which 16 are newly-sequenced species. We datamined additional 49 mosquito mitogenomes, and together with our 37 mitogenomes, we reconstructed the evolutionary history of 86 species including representatives from 15 genera and 7 tribes. Our results showed that most of the species clustered in clades with other members of their own genus with exception of Aedes genus which was paraphyletic. We confirmed the monophyletic status of the Mansoniini tribe including both Coquillettidia and Mansonia genus. The Aedeomyiini and Uranotaeniini were consistently recovered as basal to other tribes in the subfamily Culicinae, although the exact relationships among these tribes differed between analyses. These results demonstrate that low-coverage sequencing is effective to recover mitogenomes, establish phylogenetic knowledge and hence generate basic fundamental information that will help in the understanding of the role of these species as pathogen vectors.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Molaei G, Andreadis TG, Armstrong PM, Diuk-Wasser M. Host-feeding patterns of potential mosquito vectors in Connecticut, USA: molecular analysis of bloodmeals from 23 species of Aedes, Anopheles, Culex, Coquillettidia, Psorophora, and Uranotaenia. J. Med. Entomol. 2008;45:1143–1151. doi: 10.1093/jmedent/45.6.1143. - DOI - PubMed
-
- Tiawsirisup S, Nithiuthai S. Vector competence of Aedes aegypti (L.) and Culex quinquefasciatus (Say) for Dirofilaria immitis (Leidy) Southeast Asian J. Trop. Med. Public Health. 2006;37(Suppl 3):110–114. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

