Subdividing Y-chromosome haplogroup R1a1 reveals Norse Viking dispersal lineages in Britain
- PMID: 33139852
- PMCID: PMC7940619
- DOI: 10.1038/s41431-020-00747-z
Subdividing Y-chromosome haplogroup R1a1 reveals Norse Viking dispersal lineages in Britain
Abstract
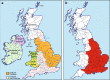
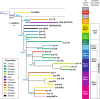
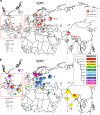
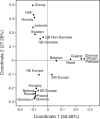
The influence of Viking-Age migrants to the British Isles is obvious in archaeological and place-names evidence, but their demographic impact has been unclear. Autosomal genetic analyses support Norse Viking contributions to parts of Britain, but show no signal corresponding to the Danelaw, the region under Scandinavian administrative control from the ninth to eleventh centuries. Y-chromosome haplogroup R1a1 has been considered as a possible marker for Viking migrations because of its high frequency in peninsular Scandinavia (Norway and Sweden). Here we select ten Y-SNPs to discriminate informatively among hg R1a1 sub-haplogroups in Europe, analyse these in 619 hg R1a1 Y chromosomes including 163 from the British Isles, and also type 23 short-tandem repeats (Y-STRs) to assess internal diversity. We find three specifically Western-European sub-haplogroups, two of which predominate in Norway and Sweden, and are also found in Britain; star-like features in the STR networks of these lineages indicate histories of expansion. We ask whether geographical distributions of hg R1a1 overall, and of the two sub-lineages in particular, correlate with regions of Scandinavian influence within Britain. Neither shows any frequency difference between regions that have higher (≥10%) or lower autosomal contributions from Norway and Sweden, but both are significantly overrepresented in the region corresponding to the Danelaw. These differences between autosomal and Y-chromosomal histories suggest either male-specific contribution, or the influence of patrilocality. Comparison of modern DNA with recently available ancient DNA data supports the interpretation that two sub-lineages of hg R1a1 spread with the Vikings from peninsular Scandinavia.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Carroll J, Harrison SH, Williams G. The Vikings in Britain and Ireland. London: British Museum Press; 2014.
-
- Abrams L, Parsons DN. Place-names and the history of Scandinavian settlement in England. In: Hines J, Lane A, Redknap M, editors. Land, sea and home: settlement in the Viking period. Leeds: Maney; 2004. p. 379–431.
-
- Kershaw J. Viking identities: Scandinavian jewellery in England. Oxford: Oxford University Press; 2013.
-
- Kershaw J, Røyrvik EC. The ‘People of the British Isles’ project and Viking settlement in England. Antiquity. 2016;90:1670–80. doi: 10.15184/aqy.2016.193. - DOI
-
- Carroll J. Identifying migrants in medieval England: the possibilities and limitations of place-name evidence. In: Ormrod WM, Tyler EM, Story J, editors. Migrants in Medieval England, c. 500–1500; proceedings of the British Academy, 219. Oxford: Oxford University Press; 2020. p. 90–119.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

