This is a preprint.
SARS-CoV-2 spike D614G variant confers enhanced replication and transmissibility
- PMID: 33140052
- PMCID: PMC7605563
- DOI: 10.1101/2020.10.27.357558
SARS-CoV-2 spike D614G variant confers enhanced replication and transmissibility
Update in
-
SARS-CoV-2 spike D614G change enhances replication and transmission.Nature. 2021 Apr;592(7852):122-127. doi: 10.1038/s41586-021-03361-1. Epub 2021 Feb 26. Nature. 2021. PMID: 33636719
Abstract
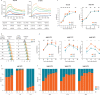
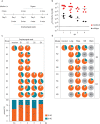
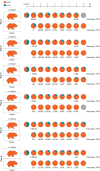
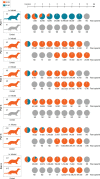
During the evolution of SARS-CoV-2 in humans a D614G substitution in the spike (S) protein emerged and became the predominant circulating variant (S-614G) of the COVID-19 pandemic 1 . However, whether the increasing prevalence of the S-614G variant represents a fitness advantage that improves replication and/or transmission in humans or is merely due to founder effects remains elusive. Here, we generated isogenic SARS-CoV-2 variants and demonstrate that the S-614G variant has (i) enhanced binding to human ACE2, (ii) increased replication in primary human bronchial and nasal airway epithelial cultures as well as in a novel human ACE2 knock-in mouse model, and (iii) markedly increased replication and transmissibility in hamster and ferret models of SARS-CoV-2 infection. Collectively, our data show that while the S-614G substitution results in subtle increases in binding and replication in vitro , it provides a real competitive advantage in vivo , particularly during the transmission bottle neck, providing an explanation for the global predominance of S-614G variant among the SARS-CoV-2 viruses currently circulating.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
References
-
- ECDC. COVID-19 pandemic, <https://www.ecdc.europa.eu/en/covid-19-pandemic> (2020).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
