KCNQ1 and Long QT Syndrome in 1/45 Amish: The Road From Identification to Implementation of Culturally Appropriate Precision Medicine
- PMID: 33141630
- PMCID: PMC7748050
- DOI: 10.1161/CIRCGEN.120.003133
KCNQ1 and Long QT Syndrome in 1/45 Amish: The Road From Identification to Implementation of Culturally Appropriate Precision Medicine
Abstract
Background: In population-based research exome sequencing, the path from variant discovery to return of results is not well established. Variants discovered by research exome sequencing have the potential to improve population health.
Methods: Population-based exome sequencing and agnostic ExWAS were performed 5521 Amish individuals. Additional phenotyping and in vitro studies enabled reclassification of a KCNQ1 variant from variant of unknown significance to pathogenic. Results were returned to participants in a community setting.
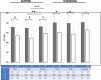
Results: A missense variant was identified in KCNQ1 (c.671C>T, p.T224M), a gene associated with long QT syndrome type 1, which can cause syncope and sudden cardiac death. The p.T224M variant, present in 1/45 Amish individuals is rare in the general population (1/248 566 in gnomAD) and was highly associated with QTc on electro-cardiogram (P=5.53E-24, β=20.2 ms/allele). Because of the potential importance of this variant to the health of the population, additional phenotyping was performed in 88 p.T224M carriers and 54 noncarriers. There was stronger clinical evidence of long QT syndrome in carriers (38.6% versus 5.5%, P=0.0006), greater history of syncope (32% versus 17%, P=0.020), and higher rate of sudden cardiac death in first degree relatives<age 30 (4.5% versus 0%, P=0.026). Expression of p.T224M KCNQ1 in Chinese hamster ovary cells showed near complete loss of protein function. Our clinical and functional data enabled reclassification of p.T224M from a variant of unknown significance to pathogenic. Of the 88 carriers, 93% met criteria for beta-blocker treatment and 5/88 (5.7%) were on medications that may further prolong QTc. Carriers were provided a Clinical Laboratory Improvement Amendments confirmed report, genetic counseling, and treatment recommendations. Follow-up care was coordinated with local physicians.
Conclusions: This work provides a framework by which research exome sequencing can be rapidly translated in a culturally appropriate manner to directly benefit research participants and enable population precision health.
Keywords: exome; genetic counseling; human; population health; syncope.
Conflict of interest statement
Drs Shuldiner, Van Hout, Gosalia, Gonzaga-Jauregu, and Economides are employees of Regeneron Pharmaceuticals, Inc and receive compensation for their employment. The other authors report no conflicts. Details are present in the
Figures




References
-
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, et al. ; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73 - PMC - PubMed
-
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Herman GE, Hufnagel SB, Klein TE, Korf BR, et al. . Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical genetics and genomics. Genet Med. 2017;19:249–255. doi: 10.1038/gim.2016.190 - PubMed
-
- Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, O’Dushlaine C, Van Hout CV, Staples J, Gonzaga-Jauregui C, et al. . Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. doi: 10.1126/science.aaf6814 - PubMed
-
- Fabsitz RR, McGuire A, Sharp RR, Puggal M, Beskow LM, Biesecker LG, Bookman E, Burke W, Burchard EG, Church G, et al. . Ethical and practical guidelines for reporting genetic research results to study participants:updated guidelines from a national heart, lung and blood institute working group. Circ Cardiovasc Genet. 2010;3:574–580. doi: 10.1161/CIRCGENETICS.110.958827 - PMC - PubMed
-
- Bombard Y, Brothers KB, Fitzgerald-Butt S, Garrison NA, Jamal L, James CA, Jarvik GP, McCormick JB, Nelson TN, Ormond KE, et al. . The responsibility to recontact research participants after reinterpretation of genetic and genomic research results. Am J Hum Genet. 2019;104:578–595. doi: 10.1016/j.ajhg.2019.02.025 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

