Emerging roles of long noncoding RNAs in cholangiocarcinoma: Advances and challenges
- PMID: 33142045
- PMCID: PMC7743012
- DOI: 10.1002/cac2.12109
Emerging roles of long noncoding RNAs in cholangiocarcinoma: Advances and challenges
Abstract
Cholangiocarcinoma (CCA), a cancer with a relatively low incidence rate, is usually associated with poor prognosis. Current modalities for the diagnosis and treatment of CCA patients are still far from satisfactory. In recent years, numerous long noncoding RNAs (lncRNAs) have been identified as crucial players in the development of various cancers, including CCA. Abnormally expressed lncRNAs in CCA, regulated by some upstream molecules, significantly influence the biological behavior of tumor cells and are involved in tumor development through various mechanisms, including interactions with functional proteins, participation in competing for endogenous RNA (ceRNA) regulatory networks, activation of cancer-related signaling pathways and epigenetic modification of gene expression. Furthermore, several lncRNAs are closely associated with the clinicopathological features of CCA patients, and are promising biomarkers for diagnosing and prognostication of CCA. Some of these lncRNAs play an important role in chemotherapy drug resistance. In addition, lncRNAs have also been shown to be involved in the inflammation microenvironment of CCA and malignant outcome of CCA risk factors, such as cholestatic liver diseases. In view of the difficulty of diagnosing CCA, more attention should be paid to detectable lncRNAs in the serum or bile. This review summarizes the recent knowledge on lncRNAs in CCA and provides a new outlook on the molecular mechanisms of CCA development from the perspective of lncRNAs. Moreover, we also discussed the limitations of the current studies and differential expression of lncRNAs in different types of CCA.
Keywords: bile; biomarkers; cancer development; cholangiocarcinoma; long noncoding RNA; molecular mechanisms; signaling pathways; tumor microenvironment.
© 2020 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
None.
Figures


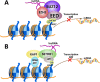
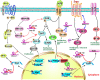

References
-
- Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861‐74. - PubMed
-
- Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, et al. DD3(PCA3)‐based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44(1):8‐16; discussion ‐6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

