Genetic and antigenic heterogeneity of GI-1/Massachusetts lineage infectious bronchitis virus variants recently isolated in China
- PMID: 33142461
- PMCID: PMC7462518
- DOI: 10.1016/j.psj.2020.08.037
Genetic and antigenic heterogeneity of GI-1/Massachusetts lineage infectious bronchitis virus variants recently isolated in China
Abstract
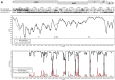
Four GI-1/Massachusetts-type (GI-1/Mass-type) infectious bronchitis virus (IBV) strains were isolated and the complete genomes of these isolates, coupled with the Mass-type live-attenuated vaccine H120 and the Mass-type pathogenic M41 strains, were sequenced in the present study. Our results show that isolates LJL/140820 and I0306/17 may be derived from the Ma5 (another Mass-type live-attenuated vaccine strain) and H120 vaccine strains, respectively. The I1124/16 strain was found to be a M41 variant that likely resulted from nucleotide accumulated mutations in the genome. Consistently, the results of the virus neutralization test showed that isolate I1124/16 was antigenically related but slight different from the M41. Our results from the protection experiments pointed out that chickens immunized with H120 failed to eliminate viral shedding after infection with the isolate I1124/16, which was different from that of M41; this result was consistent to the field observation and further implicated that the variant IBV isolate I1124/16 was antigenic different from the M41 strain. Furthermore, the I1124/16 was found to have comparable but slightly lower pathogenicity with the M41 strain. More studies based on the reverse genetic techniques are needed to elucidate the amino acids in the S1 subunit of spike protein contributing to the altered antigenicity of the isolate I1124/16. In addition, an IBV isolate, LJL/130609, was found to be originated from recombination events between the I1124/16- and Connecticut-like strains. Our results from the virus neutralization test also showed that isolates LJL/130609 and I1124/16 were antigenic closely related. Hence, there are at least 3 different genetic evolution patterns for the circulation of the GI-1/Mass-type IBV field strains in China. The differences of vaccines used, the field conditions and genetic pressures between different flocks, likely account for the emergence, evolution patterns, and characteristics of the Mass-type IBV strains.
Keywords: GI-1/Massachusetts type; antigenicity; evolution patterns; infectious bronchitis virus; pathogenicity.
Copyright © 2020. Published by Elsevier Inc.
Figures


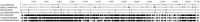


Similar articles
-
Multiple recombination events between field and vaccine strains resulted in the emergence of a novel infectious bronchitis virus with decreased pathogenicity and altered replication capacity.Poult Sci. 2020 Apr;99(4):1928-1938. doi: 10.1016/j.psj.2019.11.056. Epub 2020 Feb 28. Poult Sci. 2020. PMID: 32241473 Free PMC article.
-
Molecular and antigenic characteristics of Massachusetts genotype infectious bronchitis coronavirus in China.Vet Microbiol. 2015 Dec 31;181(3-4):241-51. doi: 10.1016/j.vetmic.2015.10.003. Epub 2015 Oct 21. Vet Microbiol. 2015. PMID: 26482289 Free PMC article.
-
Characterization of the complete genome, antigenicity, pathogenicity, tissue tropism, and shedding of a recombinant avian infectious bronchitis virus with a ck/CH/LJL/140901-like backbone and an S2 fragment from a 4/91-like virus.Virus Res. 2018 Jan 15;244:99-109. doi: 10.1016/j.virusres.2017.11.007. Epub 2017 Nov 12. Virus Res. 2018. PMID: 29141204 Free PMC article.
-
Lessons learnt on infectious bronchitis virus lineage GI-23.Avian Pathol. 2025 Feb;54(1):27-39. doi: 10.1080/03079457.2024.2398030. Epub 2024 Sep 16. Avian Pathol. 2025. PMID: 39190026 Review.
-
Avian infectious bronchitis virus.Rev Sci Tech. 2000 Aug;19(2):493-508. doi: 10.20506/rst.19.2.1228. Rev Sci Tech. 2000. PMID: 10935276 Review.
Cited by
-
Design and Characterization of a DNA Vaccine Based on Spike with Consensus Nucleotide Sequence against Infectious Bronchitis Virus.Vaccines (Basel). 2021 Jan 14;9(1):50. doi: 10.3390/vaccines9010050. Vaccines (Basel). 2021. PMID: 33466810 Free PMC article.
-
Rapid reconstruction of infectious bronchitis virus expressing fluorescent protein from its nsp2 gene based on transformation-associated recombination platform.J Virol. 2025 Jul 22;99(7):e0053525. doi: 10.1128/jvi.00535-25. Epub 2025 Jun 5. J Virol. 2025. PMID: 40470960 Free PMC article.
References
-
- Aleuy O.A., Pitesky M., Gallardo R. Using multinomial and space-time permutation models to understand the epidemiology of infectious bronchitis in California between 2008 and 2012. Avian Dis. 2018;62:226–232. - PubMed
-
- Alexander D.J., Gough R.E., Pattison M. A long-term study of the pathogenesis of infection of fowls with three strains of avian infectious bronchitis virus. Res. Vet. Sci. 1978;24:228–233. - PubMed
-
- Amarasinghe A., De Silva Senapathi U., Abdul-Cader M.S., Popowich S., Marshall F., Cork S.C., van der Meer F., Gomis S., Abdul-Careem M.F. Comparative features of infections of two Massachusetts (Mass) infectious bronchitis virus (IBV) variants isolated from Western Canadian layer flocks. BMC Vet. Res. 2018;14:391. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

