HCV Genetic Diversity Can Be Used to Infer Infection Recency and Time since Infection
- PMID: 33142675
- PMCID: PMC7692400
- DOI: 10.3390/v12111241
HCV Genetic Diversity Can Be Used to Infer Infection Recency and Time since Infection
Abstract
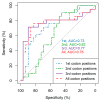
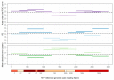
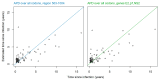
HIV-1 genetic diversity can be used to infer time since infection (TSI) and infection recency. We adapted this approach for HCV and identified genomic regions with informative diversity. We included 72 HCV/HIV-1 coinfected participants of the Swiss HIV Cohort Study, for whom reliable estimates of infection date and viral sequences were available. Average pairwise diversity (APD) was calculated over each codon position for the entire open reading frame of HCV. Utilizing cross validation, we evaluated the correlation of APD with TSI, and its ability to infer TSI via a linear model. We additionally studied the ability of diversity to classify infections as recent (infected for <1 year) or chronic, using receiver-operator-characteristic area under the curve (ROC-AUC) in 50 patients whose infection could be unambiguously classified as either recent or chronic. Measuring HCV diversity over third or all codon positions gave similar performances, and notable improvement over first or second codon positions. APD calculated over the entire genome enabled classification of infection recency (ROC-AUC = 0.76). Additionally, APD correlated with TSI (R2 = 0.33) and could predict TSI (mean absolute error = 1.67 years). Restricting the region over which APD was calculated to E2-NS2 further improved accuracy (ROC-AUC = 0.85, R2 = 0.54, mean absolute error = 1.38 years). Genetic diversity in HCV correlates with TSI and is a proxy for infection recency and TSI, even several years post-infection.
Keywords: genetic variation; hepatitis C virus infection; infection recency; sequence analysis; viral genomics.
Conflict of interest statement
HFG has received unrestricted research grants from Gilead Sciences and Roche; fees for data and safety monitoring board membership from Merck; and consulting/advisory board membership fees from Gilead Sciences, Sandoz and Mepha. EB has received fees for his institution for participation to advisory board from MSD, Gilead Sciences, ViiV Healthcare, Sandoz, Pfizer, Abbvie and Janssen. MC has received research and travel grants for his institution from ViiV and Gilead. AR reports support to his institution for advisory boards and/or travel grants from Janssen-Cilag, MSD, Gilead Sciences, Abbvie, and Bristol-Myers Squibb, and an unrestricted research grant from Gilead Sciences. All remuneration went to his home institution and not to AR personally, and all remuneration was provided outside the submitted work. KJM has received travel grants and honoraria from Gilead Sciences, Roche Diagnostics, GlaxoSmithKline, Merck Sharp & Dohme, Bristol-Myers Squibb, ViiV and Abbott; and the University of Zurich received research grants from Gilead Science, Roche, and Merck Sharp & Dohme for studies that Metzner serves as principal investigator, and advisory board honoraria from Gilead Sciences. RDK has received honoraria from Gilead Sciences (unrelated to the current work). JB received fees for his institution from Roche Glycart AG for consulting unrelated to the current work. DLB has received consulting/advisory board honoraria from Gilead Sciences, ViiV, and Merck Sharp & Dohme. MS received educational grants from Janssen-Cilag, MSD and Gilead, and Advisory Board fees from MSD, Gilead, AbbVie, ViiV, Sandoz and Mepha.
Figures

Similar articles
-
Characterization of Hepatitis C Virus (HCV) Envelope Diversification from Acute to Chronic Infection within a Sexually Transmitted HCV Cluster by Using Single-Molecule, Real-Time Sequencing.J Virol. 2017 Feb 28;91(6):e02262-16. doi: 10.1128/JVI.02262-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077634 Free PMC article.
-
A systematic, deep sequencing-based methodology for identification of mixed-genotype hepatitis C virus infections.Infect Genet Evol. 2019 Apr;69:76-84. doi: 10.1016/j.meegid.2019.01.016. Epub 2019 Jan 14. Infect Genet Evol. 2019. PMID: 30654177
-
Vertical Transmission of Hepatitis C Virus: Variable Transmission Bottleneck and Evidence of Midgestation In Utero Infection.J Virol. 2017 Nov 14;91(23):e01372-17. doi: 10.1128/JVI.01372-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28931691 Free PMC article.
-
Virological Mechanisms in the Coinfection between HIV and HCV.Mediators Inflamm. 2015;2015:320532. doi: 10.1155/2015/320532. Epub 2015 Oct 1. Mediators Inflamm. 2015. PMID: 26494946 Free PMC article. Review.
-
Coinfection of Schistosoma Species with Hepatitis B or Hepatitis C Viruses.Adv Parasitol. 2016;91:111-231. doi: 10.1016/bs.apar.2015.12.003. Epub 2016 Feb 5. Adv Parasitol. 2016. PMID: 27015949 Review.
Cited by
-
Genomic surveillance uncovers regional variation in HCV transmission networks among people who use drugs in rural U.S. communities.Res Sq [Preprint]. 2025 Jun 10:rs.3.rs-6810633. doi: 10.21203/rs.3.rs-6810633/v1. Res Sq. 2025. PMID: 40585240 Free PMC article. Preprint.
-
Human Immunodeficiency Virus (HIV) Genetic Diversity Informs Stage of HIV-1 Infection Among Patients Receiving Antiretroviral Therapy in Botswana.J Infect Dis. 2022 Apr 19;225(8):1330-1338. doi: 10.1093/infdis/jiab293. J Infect Dis. 2022. PMID: 34077517 Free PMC article.
References
-
- Cotte L., Cua E., Reynes J., Raffi F., Rey D., Delobel P., Gagneux-Brunon A., Jacomet C., Palich R., Laroche H., et al. Hepatitis C virus incidence in HIV-infected and in preexposure prophylaxis (PrEP)-using men having sex with men. Liver Int. 2018;38:1736–1740. doi: 10.1111/liv.13922. - DOI - PubMed
-
- Ghosn J., Deveau C., Goujard C., Garrigue I., Saichi N., Galimand J., Nagy Z., Rouzioux C., Meyer L., Chaix M.-L., et al. Increase in hepatitis C virus incidence in HIV-1-infected patients followed up since primary infection. Sex. Transm. Infect. 2006;82:458–460. doi: 10.1136/sti.2006.021493. - DOI - PMC - PubMed
-
- Giuliani M., Caprilli F., Gentili G., Maini A., Lepri A.C., Prignano G., Palamara G., Giglio A., Crescimbeni E., Rezza G. Incidence and Determinants of Hepatitis C Virus Infection Among Individuals at Risk of Sexually Transmitted Diseases Attending a Human Immunodeficiency Virus Type 1 Testing Program. Sex. Transm. Dis. 1997;24:533–537. doi: 10.1097/00007435-199710000-00007. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

