Snake W Sex Chromosome: The Shadow of Ancestral Amniote Super-Sex Chromosome
- PMID: 33142713
- PMCID: PMC7692289
- DOI: 10.3390/cells9112386
Snake W Sex Chromosome: The Shadow of Ancestral Amniote Super-Sex Chromosome
Abstract
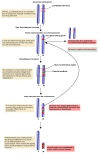
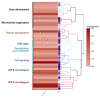
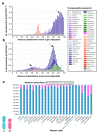
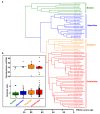
: Heteromorphic sex chromosomes, particularly the ZZ/ZW sex chromosome system of birds and some reptiles, undergo evolutionary dynamics distinct from those of autosomes. The W sex chromosome is a unique karyological member of this heteromorphic pair, which has been extensively studied in snakes to explore the origin, evolution, and genetic diversity of amniote sex chromosomes. The snake W sex chromosome offers a fascinating model system to elucidate ancestral trajectories that have resulted in genetic divergence of amniote sex chromosomes. Although the principal mechanism driving evolution of the amniote sex chromosome remains obscure, an emerging hypothesis, supported by studies of W sex chromosomes of squamate reptiles and snakes, suggests that sex chromosomes share varied genomic blocks across several amniote lineages. This implies the possible split of an ancestral super-sex chromosome via chromosomal rearrangements. We review the major findings pertaining to sex chromosomal profiles in amniotes and discuss the evolution of an ancestral super-sex chromosome by collating recent evidence sourced mainly from the snake W sex chromosome analysis. We highlight the role of repeat-mediated sex chromosome conformation and present a genomic landscape of snake Z and W chromosomes, which reveals the relative abundance of major repeats, and identifies the expansion of certain transposable elements. The latest revolution in chromosomics, i.e., complete telomere-to-telomere assembly, offers mechanistic insights into the evolutionary origin of sex chromosomes.
Keywords: chromosomal rearrangements; evolution; genome; next-generation sequencing; repeat elements; sex determination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an "ancestral amniote super-sex chromosome"?Chromosome Res. 2020 Jun;28(2):209-228. doi: 10.1007/s10577-020-09631-4. Epub 2020 May 1. Chromosome Res. 2020. PMID: 32358743
-
Chromosome map of the Siamese cobra: did partial synteny of sex chromosomes in the amniote represent "a hypothetical ancestral super-sex chromosome" or random distribution?BMC Genomics. 2018 Dec 17;19(1):939. doi: 10.1186/s12864-018-5293-6. BMC Genomics. 2018. PMID: 30558533 Free PMC article.
-
Partial Amniote Sex Chromosomal Linkage Homologies Shared on Snake W Sex Chromosomes Support the Ancestral Super-Sex Chromosome Evolution in Amniotes.Front Genet. 2020 Aug 18;11:948. doi: 10.3389/fgene.2020.00948. eCollection 2020. Front Genet. 2020. PMID: 33014016 Free PMC article.
-
The Diversity and Evolution of Sex Chromosomes in Frogs.Genes (Basel). 2021 Mar 26;12(4):483. doi: 10.3390/genes12040483. Genes (Basel). 2021. PMID: 33810524 Free PMC article. Review.
-
Turtle Insights into the Evolution of the Reptilian Karyotype and the Genomic Architecture of Sex Determination.Genes (Basel). 2020 Apr 11;11(4):416. doi: 10.3390/genes11040416. Genes (Basel). 2020. PMID: 32290488 Free PMC article. Review.
Cited by
-
Why Do Some Vertebrates Have Microchromosomes?Cells. 2021 Aug 24;10(9):2182. doi: 10.3390/cells10092182. Cells. 2021. PMID: 34571831 Free PMC article. Review.
-
Sex chromosome evolution among amniotes: is the origin of sex chromosomes non-random?Philos Trans R Soc Lond B Biol Sci. 2021 Sep 13;376(1833):20200108. doi: 10.1098/rstb.2020.0108. Epub 2021 Jul 26. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34304592 Free PMC article.
-
Insights into avian molecular cytogenetics-with reptilian comparisons.Mol Cytogenet. 2024 Oct 31;17(1):24. doi: 10.1186/s13039-024-00696-y. Mol Cytogenet. 2024. PMID: 39482771 Free PMC article. Review.
-
The Rattlesnake W Chromosome: A GC-Rich Retroelement Refugium with Retained Gene Function Across Ancient Evolutionary Strata.Genome Biol Evol. 2022 Sep 6;14(9):evac116. doi: 10.1093/gbe/evac116. Genome Biol Evol. 2022. PMID: 35867356 Free PMC article.
-
Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation?Life (Basel). 2022 Apr 1;12(4):522. doi: 10.3390/life12040522. Life (Basel). 2022. PMID: 35455013 Free PMC article. Review.
References
-
- Hake L., O’Connor C. Genetic mechanisms of sex determination. Nature. 2008;1:25.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

