CancerGram: An Effective Classifier for Differentiating Anticancer from Antimicrobial Peptides
- PMID: 33142753
- PMCID: PMC7692641
- DOI: 10.3390/pharmaceutics12111045
CancerGram: An Effective Classifier for Differentiating Anticancer from Antimicrobial Peptides
Abstract
Antimicrobial peptides (AMPs) constitute a diverse group of bioactive molecules that provide multicellular organisms with protection against microorganisms, and microorganisms with weaponry for competition. Some AMPs can target cancer cells; thus, they are called anticancer peptides (ACPs). Due to their small size, positive charge, hydrophobicity and amphipathicity, AMPs and ACPs interact with negatively charged components of biological membranes. AMPs preferentially permeabilize microbial membranes, but ACPs additionally target mitochondrial and plasma membranes of cancer cells. The preference towards mitochondrial membranes is explained by their membrane potential, membrane composition resulting from α-proteobacterial origin and the fact that mitochondrial targeting signals could have evolved from AMPs. Taking into account the therapeutic potential of ACPs and millions of deaths due to cancer annually, it is of vital importance to find new cationic peptides that selectively destroy cancer cells. Therefore, to reduce the costs of experimental research, we have created a robust computational tool, CancerGram, that uses n-grams and random forests for predicting ACPs. Compared to other ACP classifiers, CancerGram is the first three-class model that effectively classifies peptides into: ACPs, AMPs and non-ACPs/non-AMPs, with AU1U amounting to 0.89 and a Kappa statistic of 0.65. CancerGram is available as a web server and R package on GitHub.
Keywords: anticancer peptide (ACP); anticancer peptides; antimicrobial peptide (AMP); antimicrobial peptides; host defense peptides; prediction; random forest.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

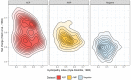
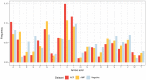
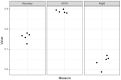
Similar articles
-
Proteomic Screening for Prediction and Design of Antimicrobial Peptides with AmpGram.Int J Mol Sci. 2020 Jun 17;21(12):4310. doi: 10.3390/ijms21124310. Int J Mol Sci. 2020. PMID: 32560350 Free PMC article.
-
Antimicrobial/anticancer peptides: bioactive molecules and therapeutic agents.Immunotherapy. 2021 Jun;13(8):669-684. doi: 10.2217/imt-2020-0312. Epub 2021 Apr 21. Immunotherapy. 2021. PMID: 33878901 Review.
-
Exploratory data analysis of physicochemical parameters of natural antimicrobial and anticancer peptides: Unraveling the patterns and trends for the rational design of novel peptides.Bioimpacts. 2024;14(1):26438. doi: 10.34172/bi.2023.26438. Epub 2023 Aug 20. Bioimpacts. 2024. PMID: 38327633 Free PMC article.
-
Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities.Molecules. 2020 Jun 19;25(12):2850. doi: 10.3390/molecules25122850. Molecules. 2020. PMID: 32575664 Free PMC article. Review.
-
Incorporating support vector machine with sequential minimal optimization to identify anticancer peptides.BMC Bioinformatics. 2021 May 29;22(1):286. doi: 10.1186/s12859-021-03965-4. BMC Bioinformatics. 2021. PMID: 34051755 Free PMC article.
Cited by
-
Glioblastoma Multiforme: Sensitivity to Antimicrobial Peptides LL-37 and PG-1, and Their Combination with Chemotherapy for Predicting the Overall Survival of Patients.Pharmaceutics. 2024 Sep 22;16(9):1234. doi: 10.3390/pharmaceutics16091234. Pharmaceutics. 2024. PMID: 39339270 Free PMC article.
-
Cationic antimicrobial peptides: potential templates for anticancer agents.Front Med (Lausanne). 2025 Apr 24;12:1548603. doi: 10.3389/fmed.2025.1548603. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40342581 Free PMC article. Review.
-
Jackfruit Leaf Protein Hydrolysates Obtained by Enzymatic Hydrolysis of Leaf Protein Concentrate with Pepsin and Pancreatin: Molecular Weight, Cytotoxicity, Antiproliferative Activity, and Oxidative Stress.Plant Foods Hum Nutr. 2024 Sep;79(3):685-692. doi: 10.1007/s11130-024-01203-9. Epub 2024 Jul 10. Plant Foods Hum Nutr. 2024. PMID: 38985367
-
Lactoferricin B Combined with Antibiotics Exhibits Leukemic Selectivity and Antimicrobial Activity.Molecules. 2024 Feb 1;29(3):678. doi: 10.3390/molecules29030678. Molecules. 2024. PMID: 38338422 Free PMC article.
-
Role of Anti-Cancer Peptides as Immunomodulatory Agents: Potential and Design Strategy.Pharmaceutics. 2022 Dec 1;14(12):2686. doi: 10.3390/pharmaceutics14122686. Pharmaceutics. 2022. PMID: 36559179 Free PMC article. Review.
References
-
- Cancer. [(accessed on 13 October 2020)]; Available online: https://www.who.int/news-room/fact-sheets/detail/cancer.
-
- Cassini A., Högberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
-
- CDC . Antibiotic Resistance Threats in the United States, 2019. Centres for Disease Control and Prevention, US Department of Health and Human Services; Washington, DC, USA: 2019.
Grants and funding
LinkOut - more resources
Full Text Sources

