Origin and Evolution of Studiervirinae Bacteriophages Infecting Pectobacterium: Horizontal Transfer Assists Adaptation to New Niches
- PMID: 33142811
- PMCID: PMC7693777
- DOI: 10.3390/microorganisms8111707
Origin and Evolution of Studiervirinae Bacteriophages Infecting Pectobacterium: Horizontal Transfer Assists Adaptation to New Niches
Abstract
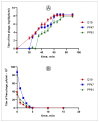
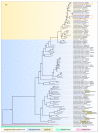
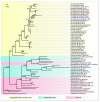
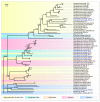
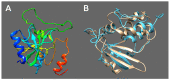
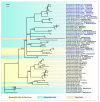
Black leg and soft rot are devastating diseases causing up to 50% loss of potential potato yield. The search for, and characterization of, bacterial viruses (bacteriophages) suitable for the control of these diseases is currently a sought-after task for agricultural microbiology. Isolated lytic Pectobacterium bacteriophages Q19, PP47 and PP81 possess a similar broad host range but differ in their genomic properties. The genomic features of characterized phages have been described and compared to other Studiervirinae bacteriophages. Thorough phylogenetic analysis has clarified the taxonomy of the phages and their positioning relative to other genera of the Autographiviridae family. Pectobacterium phage Q19 seems to represent a new genus not described previously. The genomes of the phages are generally similar to the genome of phage T7 of the Teseptimavirus genus but possess a number of specific features. Examination of the structure of the genes and proteins of the phages, including the tail spike protein, underlines the important role of horizontal gene exchange in the evolution of these phages, assisting their adaptation to Pectobacterium hosts. The results provide the basis for the development of bacteriophage-based biocontrol of potato soft rot as an alternative to the use of antibiotics.
Keywords: Autographiviridae; Pectobacterium; evolution; horizontal transfer; phage; phage therapy; tail spike.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures








References
-
- Czajkowski R., Pérombelon M.C.M., Van Veen J.A., Van der Wolf J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant. Pathol. 2011;60:999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. - DOI
-
- Pérombelon M.C.M. Potato diseases caused by soft rot erwinias: An overview of pathogenesis. Plant. Pathol. 2002;51:1–12.
-
- Ma B., Hibbing M.E., Kim H.-S., Reedy R.M., Yedidia I., Breuer J.J., Breuer J.J., Glasner J.D., Perna N.T., Kelman A., et al. Host range and molecular phylogenies of the soft rot enterobacterial genera pectobacterium and dickeya. Phytopathology. 2007;97:1150–1163. doi: 10.1094/PHYTO-97-9-1150. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

