Yeast Rpn4 Links the Proteasome and DNA Repair via RAD52 Regulation
- PMID: 33143019
- PMCID: PMC7672625
- DOI: 10.3390/ijms21218097
Yeast Rpn4 Links the Proteasome and DNA Repair via RAD52 Regulation
Abstract
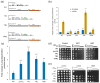
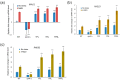
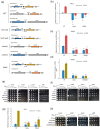
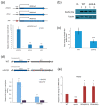
Environmental and intracellular factors often damage DNA, but multiple DNA repair pathways maintain genome integrity. In yeast, the 26S proteasome and its transcriptional regulator and substrate Rpn4 are involved in DNA damage resistance. Paradoxically, while proteasome dysfunction may induce hyper-resistance to DNA-damaging agents, Rpn4 malfunction sensitizes yeasts to these agents. Previously, we proposed that proteasome inhibition causes Rpn4 stabilization followed by the upregulation of Rpn4-dependent DNA repair genes and pathways. Here, we aimed to elucidate the key Rpn4 targets responsible for DNA damage hyper-resistance in proteasome mutants. We impaired the Rpn4-mediated regulation of candidate genes using the CRISPR/Cas9 system and tested the sensitivity of mutant strains to 4-NQO, MMS and zeocin. We found that the separate or simultaneous deregulation of 19S or 20S proteasome subcomplexes induced MAG1, DDI1, RAD23 and RAD52 in an Rpn4-dependent manner. Deregulation of RAD23, DDI1 and RAD52 sensitized yeast to DNA damage. Genetic, epigenetic or dihydrocoumarin-mediated RAD52 repression restored the sensitivity of the proteasome mutants to DNA damage. Our results suggest that the Rpn4-mediated overexpression of DNA repair genes, especially RAD52, defines the DNA damage hyper-resistant phenotype of proteasome mutants. The developed yeast model is useful for characterizing drugs that reverse the DNA damage hyper-resistance phenotypes of cancers.
Keywords: 26S proteasome; CRISPR/Cas9; DDI1; DNA repair; RAD23; RAD52; Rpn4.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Proteasome inhibition enhances resistance to DNA damage via upregulation of Rpn4-dependent DNA repair genes.FEBS Lett. 2013 Sep 17;587(18):3108-14. doi: 10.1016/j.febslet.2013.08.007. Epub 2013 Aug 13. FEBS Lett. 2013. PMID: 23954292
-
Rpn4 and proteasome-mediated yeast resistance to ethanol includes regulation of autophagy.Appl Microbiol Biotechnol. 2020 May;104(9):4027-4041. doi: 10.1007/s00253-020-10518-x. Epub 2020 Mar 10. Appl Microbiol Biotechnol. 2020. PMID: 32157425
-
Inhibition of proteasomal degradation of rpn4 impairs nonhomologous end-joining repair of DNA double-strand breaks.PLoS One. 2010 Apr 1;5(4):e9877. doi: 10.1371/journal.pone.0009877. PLoS One. 2010. PMID: 20376190 Free PMC article.
-
Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function.Biochim Biophys Acta. 2007 Nov;1773(11):1599-604. doi: 10.1016/j.bbamcr.2007.05.015. Epub 2007 Jun 12. Biochim Biophys Acta. 2007. PMID: 17604855 Review.
-
A role for Rad23 proteins in 26S proteasome-dependent protein degradation?Mutat Res. 2002 Jan 29;499(1):53-61. doi: 10.1016/s0027-5107(01)00291-3. Mutat Res. 2002. PMID: 11804604 Review.
Cited by
-
Tissue-specific DamID protocol using nanopore sequencing.J Biol Methods. 2021 Aug 27;8(3):e152. doi: 10.14440/jbm.2021.362. eCollection 2021. J Biol Methods. 2021. PMID: 34514013 Free PMC article.
-
Synergistic Effect of a Combination of Proteasome and Ribonucleotide Reductase Inhibitors in a Biochemical Model of the Yeast Saccharomyces cerevisiae and a Glioblastoma Cell Line.Int J Mol Sci. 2024 Apr 3;25(7):3977. doi: 10.3390/ijms25073977. Int J Mol Sci. 2024. PMID: 38612788 Free PMC article.
-
DNA damage and regulation of protein homeostasis.DNA Repair (Amst). 2021 Sep;105:103155. doi: 10.1016/j.dnarep.2021.103155. Epub 2021 Jun 8. DNA Repair (Amst). 2021. PMID: 34116476 Free PMC article. Review.
-
Yeast Ribonucleotide Reductase Is a Direct Target of the Proteasome and Provides Hyper Resistance to the Carcinogen 4-NQO.J Fungi (Basel). 2023 Mar 14;9(3):351. doi: 10.3390/jof9030351. J Fungi (Basel). 2023. PMID: 36983519 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

