A Review of the Important Role of CYP2D6 in Pharmacogenomics
- PMID: 33143137
- PMCID: PMC7692531
- DOI: 10.3390/genes11111295
A Review of the Important Role of CYP2D6 in Pharmacogenomics
Abstract
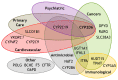
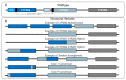
Cytochrome P450 2D6 (CYP2D6) is a critical pharmacogene involved in the metabolism of ~20% of commonly used drugs across a broad spectrum of medical disciplines including psychiatry, pain management, oncology and cardiology. Nevertheless, CYP2D6 is highly polymorphic with single-nucleotide polymorphisms, small insertions/deletions and larger structural variants including multiplications, deletions, tandem arrangements, and hybridisations with non-functional CYP2D7 pseudogenes. The frequency of these variants differs across populations, and they significantly influence the drug-metabolising enzymatic function of CYP2D6. Importantly, altered CYP2D6 function has been associated with both adverse drug reactions and reduced drug efficacy, and there is growing recognition of the clinical and economic burdens associated with suboptimal drug utilisation. To date, pharmacogenomic clinical guidelines for at least 48 CYP2D6-substrate drugs have been developed by prominent pharmacogenomics societies, which contain therapeutic recommendations based on CYP2D6-predicted categories of metaboliser phenotype. Novel algorithms to interpret CYP2D6 function from sequencing data that consider structural variants, and machine learning approaches to characterise the functional impact of novel variants, are being developed. However, CYP2D6 genotyping is yet to be implemented broadly into clinical practice, and so further effort and initiatives are required to overcome the implementation challenges and deliver the potential benefits to the bedside.
Keywords: CYP2D6; drug metabolism; pharmacogenomics; population variability; structural variation.
Conflict of interest statement
Christopher Taylor, Ian Crosby and Peter Maguire work with MC Diagnostics which operates as a commercial genetic testing kit manufacturer. The remaining authors have not conflict of interest to declare.
Figures


References
-
- Šrejber M., Navrátilová V., Paloncýová M., Bazgier V., Berka K., Anzenbacher P., Otyepka M. Membrane-attached mammalian cytochromes P450: An overview of the membrane’s effects on structure, drug binding, and interactions with redox partners. J. Inorg. Biochem. 2018;183:117–136. doi: 10.1016/j.jinorgbio.2018.03.002. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

