Achimota Pararubulavirus 3: A New Bat-Derived Paramyxovirus of the Genus Pararubulavirus
- PMID: 33143230
- PMCID: PMC7692193
- DOI: 10.3390/v12111236
Achimota Pararubulavirus 3: A New Bat-Derived Paramyxovirus of the Genus Pararubulavirus
Abstract
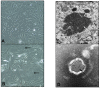
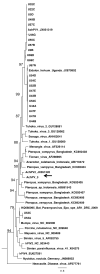
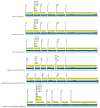
Bats are an important source of viral zoonoses, including paramyxoviruses. The paramyxoviral Pararubulavirus genus contains viruses mostly derived from bats that are common, diverse, distributed throughout the Old World, and known to be zoonotic. Here, we describe a new member of the genus Achimota pararubulavirus 3 (AchPV3) and its isolation from the urine of African straw-coloured fruit bats on primary bat kidneys cells. We sequenced and analysed the genome of AchPV3 relative to other Paramyxoviridae, revealing it to be similar to known pararubulaviruses. Phylogenetic analysis of AchPV3 revealed the failure of molecular detection in the urine sample from which AchPV3 was derived and an attachment protein most closely related with AchPV2-a pararubulavirus known to cause cross-species transmission. Together these findings add to the picture of pararubulaviruses, their sources, and variable zoonotic potential, which is key to our understanding of host restriction and spillover of bat-derived paramyxoviruses. AchPV3 represents a novel candidate zoonosis and an important tool for further study.
Keywords: bat; electron microscopy; genomics; molecular detection; paramyxovirus; pararubulavirus; primary cell lines; virus; virus discovery; zoonosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Baker K.S., Leggett R.M., Bexfield N.H., Alston M., Daly G., Todd S., Tachedjian M., Holmes C.E., Crameri S., Wang L.F., et al. Metagenomic study of the viruses of African straw-coloured fruit bats: Detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95–106. doi: 10.1016/j.virol.2013.03.014. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

