p53 Loss Mediates Hypersensitivity to ETS Transcription Factor Inhibition Based on PARylation-Mediated Cell Death Induction
- PMID: 33143299
- PMCID: PMC7693367
- DOI: 10.3390/cancers12113205
p53 Loss Mediates Hypersensitivity to ETS Transcription Factor Inhibition Based on PARylation-Mediated Cell Death Induction
Abstract
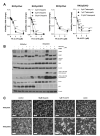
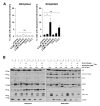
The small-molecule E26 transformation-specific (ETS) factor inhibitor YK-4-279 was developed for therapy of ETS/EWS fusion-driven Ewing's sarcoma. Here we aimed to identify molecular factors underlying YK-4-279 responsiveness in ETS fusion-negative cancers. Cell viability screenings that deletion of P53 induced hypersensitization against YK-4-279 especially in the BRAFV600E-mutated colon cancer model RKO. This effect was comparably minor in the BRAF wild-type HCT116 colon cancer model. Out of all ETS transcription factor family members, especially ETS1 overexpression at mRNA and protein level was induced by deletion of P53 specifically under BRAF-mutated conditions. Exposure to YK-4-279 reverted ETS1 upregulation induced by P53 knock-out in RKO cells. Despite upregulation of p53 by YK-4-279 itself in RKOp53 wild-type cells, YK-4-279-mediated hyperphosphorylation of histone histone H2A.x was distinctly more pronounced in the P53 knock-out background. YK-4-279-induced cell death in RKOp53-knock-out cells involved hyperPARylation of PARP1, translocation of the apoptosis-inducible factor AIF into nuclei, and induction of mitochondrial membrane depolarization, all hallmarks of parthanatos. Accordingly, pharmacological PARP as well as BRAFV600E inhibition showed antagonistic activity with YK-4-279 especially in the P53 knock-out background. Taken together, we identified ETS factor inhibition as a promising strategy for the treatment of notoriously therapy-resistant p53-null solid tumours with activating MAPK mutations.
Keywords: ETS factor inhibitor; ETS1; PARylation; YK-4-279; p53; parthanatos.
Conflict of interest statement
M.P. has received honoraria for lectures, consultation or advisory board participation from the following for-profit companies: Bayer, Bristol-Myers Squibb, Novartis, Gerson Lehrman Group (GLG), CMC Contrast, GlaxoSmithKline, Mundipharma, Roche, BMJ Journals, MedMedia, Astra Zeneca, AbbVie, Lilly, Medahead, Daiichi Sankyo, Sanofi, Merck Sharp & Dome, Tocagen. The following for-profit companies have supported clinical trials and contracted research conducted by MP with payments made to his institution: Böhringer-Ingelheim, Bristol-Myers Squibb, Roche, Daiichi Sankyo, Merck Sharp and Dome, Novocure, GlaxoSmithKline, AbbVie. WB has received honoraria for lectures, consultation or advisory board participation from: Bristol-Myers Squibb, Novartis, Merck Sharp & Dome, Roche, Astra Zeneca and AbbVie. All other authors declare no conflicts of interest.
Figures
References
-
- Birner P., Berghoff A.S., Dinhof C., Pirker C., Capper D., Schoppmann S.F., Petzelbauer P., von Deimling A., Berger W., Preusser M. MAP kinase activity supported by BRAF V600E mutation rather than gene amplification is associated with ETV1 expression in melanoma brain metastases. Arch. Dermatol. Res. 2014;306:873–884. doi: 10.1007/s00403-014-1490-6. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

