Aprotinin Inhibits SARS-CoV-2 Replication
- PMID: 33143316
- PMCID: PMC7692688
- DOI: 10.3390/cells9112377
Aprotinin Inhibits SARS-CoV-2 Replication
Abstract
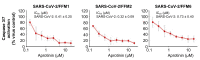
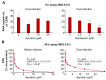
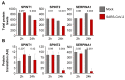
Severe acute respiratory syndrome virus 2 (SARS-CoV-2) is the cause of the current coronavirus disease 19 (COVID-19) pandemic. Protease inhibitors are under consideration as virus entry inhibitors that prevent the cleavage of the coronavirus spike (S) protein by cellular proteases. Herein, we showed that the protease inhibitor aprotinin (but not the protease inhibitor SERPINA1/alpha-1 antitrypsin) inhibited SARS-CoV-2 replication in therapeutically achievable concentrations. An analysis of proteomics and translatome data indicated that SARS-CoV-2 replication is associated with a downregulation of host cell protease inhibitors. Hence, aprotinin may compensate for downregulated host cell proteases during later virus replication cycles. Aprotinin displayed anti-SARS-CoV-2 activity in different cell types (Caco2, Calu-3, and primary bronchial epithelial cell air-liquid interface cultures) and against four virus isolates. In conclusion, therapeutic aprotinin concentrations exert anti-SARS-CoV-2 activity. An approved aprotinin aerosol may have potential for the early local control of SARS-CoV-2 replication and the prevention of COVID-19 progression to a severe, systemic disease.
Keywords: 2019-nCoV; COVID-19; antiviral; aprotinin; drug discovery; severe acute respiratory syndrome coronavirus; severe acute respiratory syndrome coronavirus 2.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

